Document Type : Original Research Article
Authors
1 Associated Professor, dep of Lab Medical Sciences, Faculty of allied Medical Sciences, Tehran University of Medical Sciences, Tehran, Iran
2 Ph.D Microbiology, Food microbiology research center, Tehran University of Medical Sciences, Tehran, Iran
3 Department of Molecular Cell Biology-Microbiology, Faculty of Convergent Sciences and Technologies, Science and Research Branch, Islamic Azad University, Tehran, Iran
Abstract
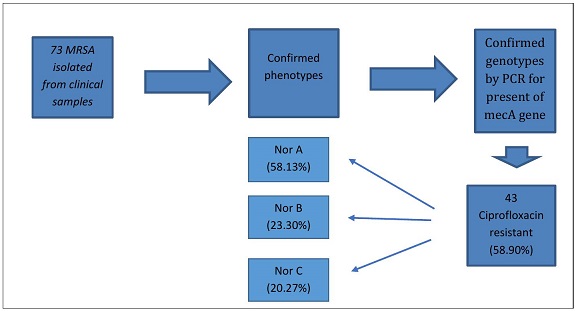
Staphylococcus aureus (S. aureus) is an important pathogen that is involved in causing various infections. The most important problem in the treatment of staphylococcal infections is the increasing drug resistance of these bacteria. In this research, the presence of efflux pump genes in methicillin-resistant Staphylococcus aureus strains isolated from clinical samples was investigated. 73 MRSA were collected and confirmed phenotypically and biochemically. PCR was performed by checking the presence of mecA, vanA, norA, norB, and norC genes. All 73 clinical samples underwent meticulous confirmation, unequivocally establishing their classification as Methicillin-Resistant Staphylococcus aureus (MRSA). Delving into the antibiotic resistance landscape, a substantial portion of the MRSA strains (58.90%) manifested resistance to ciprofloxacin, with the norA gene emerging as the predominant contributor in 25 instances (58.13%). Frequency of vanA genes among isolated MRSA was 5 (6.84%). Furthermore, a nuanced genetic complexity was unveiled as certain MRSA isolates exhibited the presence of all three norA, norB, norC genes concurrently, or alternatively, the co-occurrence of two among them. This intricate molecular profile accentuates the multifaceted nature of antibiotic resistance mechanisms within the MRSA population under scrutiny, underscoring the need for a comprehensive understanding to inform targeted therapeutic interventions. The presence of vanA and efflux pump resistance genes among the studied isolated MRSA is a warning for the spread of MDR strains in clinical isolates. This current study further underscores the peril associated with such infections within hospital systems. Given the elevated clinical significance of these infections, it becomes imperative to prioritize the awareness of healthcare professionals. Consequently, enhancing the vigilance and preparedness of medical practitioners becomes paramount in addressing the evolving landscape of antimicrobial resistance within healthcare settings.
Graphical Abstract
Keywords
Main Subjects
Introduction
Infections stemming from methicillin-resistant Staphylococcus aureus (MRSA) strains are pervasive in both societal and hospital settings, primarily attributable to Staphylococcus aureus' capacity to colonize areas like the nose or skin, subsequently precipitating disease dissemination. Staphylococci have emerged as a leading cause of hospital-acquired infections, with the surge in drug-resistant strains, particularly MRSA, posing a formidable challenge in treating and controlling these infections [1]. Initial studies indicated minimal MRSA resistance to ciprofloxacin, showcasing efficacy in treating experimental MRSA endocarditis. However, recent reports signal an escalating resistance trend [2].
The global rise in antimicrobial resistance, notably among hospital pathogens, is a contemporary concern. Multidrug-resistant Staphylococci present an escalating threat to human health [3]. The principal resistance mechanism in MRSA strains against penicillin involves beta-lactamase production, which hydrolyzes the penicillin beta-lactam ring, rendering it inactive. Another mechanism entails the penicillin-binding protein PBP2a, encoded by mecA2 [4].
The MFS efflux system, particularly the norA gene, emerges as a pivotal player in Staphylococcus aureus. Encoding a 388-amino acid protein organized into 12 transmembrane segments, NorA shares 24% similarity with the Tet (A) efflux pump in E. coli [5-6].The NorB efflux pump confers resistance to various NorA substrates, encompassing hydrophilic fluoroquinolones, biocides, and dyes. Similarly, NorC, encoded by norC, shares structural homology with NorB and is associated with low-level resistance to diverse fluoroquinolones and dyes [7-8].
There are different mechanisms in bacteria related to ciprofloxacin resistance which included mutations on gyrA, T83I, D87Y, mexZ, UmuC, qnr, and other genes. This study delves into efflux pump genes in ciprofloxacin-resistant MRSA isolates from clinical samples across diverse hospital departments affiliated with Tehran University of Medical Sciences. Emphasizing the urgency of this matter, the research sheds light on the intricacies of antibiotic resistance mechanisms, paving the way for informed interventions in the face of this evolving healthcare challenge.
Materials and methods
Collection of Clinical Samples
73 culture plates were prepared from clinical samples suspected of methicillin-resistant Staphylococcus aureus (MRSA) obtained from different departments of the hospitals affiliated to Tehran University of Medical Sciences in 2022-2023. These samples were obtained from the hospital laboratory.
Phenotypic confirmation of Staphylococcus aureus isolates
All submitted plates were isolated on Blood agar and Nutrient agar. After observing hemolysis type on Blood agar, Gram staining, catalase test, coagulase differentiation test, urease test, mannitol fermentation test, DNase test, and coagulase test were performed.
Determination of antibiotic sensitivity by the disk diffusion method
Isolates confirmed as Staphylococcus aureus were cultured on NA medium and incubated at 37 degrees Celsius for 18-24 hours. A pure colony from the fresh culture was added to 0.9% physiological saline solution to create a turbidity equivalent to half McFarland. The microbial suspension with a half McFarland turbidity was streaked on Muller-Hinton agar in three directions. After a few minutes, antibiotic discs including Amoxicillin (An), Trimethoprim-sulfamethoxazole (SXT), Cefazolin (CZ), Ceftriaxone (CRO), Ciprofloxacin (CP), Erythromycin (E), Imipenem (IPM), Cefotaxime (CTX), Clindamycin (CC), Cloxacillin (CX), Vancomycin (V), Cefoxitin (FOX), and Linezolid (LZV) were placed on Muller-Hinton agar at a distance of 24 mm from each other. The results for the discs were recorded as sensitive (S), intermediate (I), and resistant (R). Strains resistant to ciprofloxacin were further investigated for the presence of norA, norB, and norC genes (Figure 1).
Genomic DNA extraction was performed using the Boiling method:
- Bacteria were cultured in TSA medium and incubated for 24 hours at 37
- A pure colony of bacteria was dissolved in 400 microliters of sterile distilled water.
- The bacterial suspension was placed in a Thermo block device at 100 C for 20 minutes.
- The bacterial suspension was centrifuged at 13,000 rpm for 10 minutes.
- Approximately 300 microliters of the supernatant were transferred to a sterile microtube and stored in a -20-degree centigrade freezer for PCR testing.
PCR Test
For the PCR test, after obtaining the desired primers (Table 1), necessary materials for the PCR assay and under the specified temperature conditions (Table 2), the test was conducted using the PeQlab thermal cycler model peQSTAR. The PCR product was then tracked by electrophoresis on a 1% agarose gel with X5/0 TBE buffer, at a voltage of 103 volts for approximately 37 minutes. Finally, the gel image was prepared using a gel documentation system. Optimization of PCR conditions was carried out in a volume of 20 microliters.
Statistical analysis
Descriptive information was analyzed using the frequency test and determination of means. Qualitative analysis was performed using the χ2 test in SPSS software version 21. A significance level of P < 0.05 was considered for statistical significance.
Results
In this inquiry, a total of 73 samples were scrutinized, revealing a gender distribution of 42 (54.57%) from males and 31 (46.42%) from females. Phenotypic and biochemical tests definitively verified the presence of Staphylococcus aureus. To ascertain methicillin resistance and identify Methicillin-Resistant Staphylococcus aureus (MRSA), cefoxitin disks were employed. Subsequently, all strains confirmed as methicillin-resistant were unequivocally categorized as MRSA. Despite the comprehensive analysis, no statistically significant association surfaced between gender and MRSA prevalence, as indicated by a P-value of 0.1. This underscores the need for further exploration and nuanced considerations in unraveling the factors contributing to MRSA incidence, extending beyond gender-based correlations.
Prevalence of MRSA in different hospital departments
The results of the investigation into the average prevalence in various hospital departments indicate that the Intensive Care Unit (ICU) had the highest number of clinical samples suspected of MRSA, with 25 cases (24.34%), while the Ear, Nose, and Throat (ENT) department had the lowest with 1 case (1.36%). The findings are presented in Table 3.
The collected samples, categorized based on their source (site of sample acquisition), are presented in Table 4. The highest prevalence of MRSA isolates, 21 cases (77.28%), was found in wound samples, while synovial fluid exhibited the lowest prevalence with one case (1.36%). A significant correlation was observed between the prevalence of MRSA and hospital departments (P = 0.008).
Determination of Antimicrobial Sensitivity Pattern
Based on the antibiotics listed in CLSI 2022, an antibiogram was conducted using the Kirby-Bauer method. The results indicated that all strains were sensitive to Linezolid and Vancomycin (100%) and resistant to Cefoxitin (100%). The findings are presented in Table 5.
Molecular Identification of MRSA and detection of mecA, vanA, norA, norB, and norC Genes
In this investigation, a comprehensive analysis was conducted on 73 clinical isolates of Methicillin-Resistant Staphylococcus aureus (MRSA) obtained from three prominent Tehran hospitals. Polymerase Chain Reaction (PCR) was employed to scrutinize the genetic landscape associated with their epidemiological characteristics, with a particular focus on mecA, vanA, norA, norB, and norC genes. All isolates demonstrating resistance to cefoxitin underwent molecular assessment for the presence of the mecA gene, revealing that every single one harbored this key genetic marker (Figure 2). The disc diffusion method is not a suitable method for detection of vancomycin resistance strain, and in this research, the existence of gene vanA was investigated by PCR method. In addition, the investigation into vancomycin resistance in MRSA isolates, as determined by disk diffusion, unveiled that 5 isolates (6.84%) of MRSA carried the vanA gene, underscoring the prevalence of vancomycin resistance within this subset.
Among the MRSA isolates, a noteworthy 90.58% exhibited resistance to ciprofloxacin. The distribution of nor genes in ciprofloxacin-resistant samples revealed that the norA gene was present in 25 (58.13%) isolates, norB in 13 (23.30%), and norC in 12 (20.27%). Within the subset of neither ciprofloxacin-resistant isolates harboring nor genes, a striking 60.6% simultaneously carried all three norA, B, and C genes. These multifaceted isolates were sourced from diverse departments, including surgical, Neonatal Intensive Care Unit (NICU), and pediatric units, originating from samples such as urine and cerebrospinal fluid. Moreover, another set of isolates (60.6%) concurrently harbored both norA, B genes, sourced from ICU, pediatric, and dermatology departments, and originating from wound and urine samples. Similarly, a distinct group (60.6%) simultaneously carried both norB, C genes, associated with burn, surgical, and oncology departments, and sourced from blood and abscess samples. Intriguingly, the simultaneous presence of only two norA, C genes was not observed in any of the isolates. Detailed results pertaining to these genetic findings are visually depicted in Figures 3 to 6, offering a comprehensive insight into the intricate genetic profiles of the examined MRSA isolates.
Discussion
Staphylococcus aureus is one of the most significant pathogenic bacteria and one of the four major causes of nosocomial infections. The other bacterial agents included E.coli, Pseudomonas, and klebsiella. This bacterium is both toxigenic and pathogenic in humans, contributing to morbidity and mortality, especially in immunocompromised and patients with underlying conditions. Toxins produced by Staphylococcus aureus include, cytolitic toxin, epidermolytic toxin, pyrogenic toxin, and enterotoxin (12). In this study, the male population demonstrated a higher susceptibility to MRSA infection at 54.57% compared to females. The current study did not find a significant correlation between age, gender, and MRSA infection, while some studies have reported no significant association between gender and MRSA infection (13). Others have suggested a higher prevalence of this infection in young individuals, while some studies have associated it with the elderly (14). In addition, specific studies have considered men (15), especially rural men (16), to be more susceptible to MRSA infections. Thus, it can be inferred that while most studies claim no significant relationship between gender and age with MRSA infection, occasional studies suggesting a meaningful correlation may be influenced by anatomical factors, hygiene practices, environmental experiences, stress, and exposure to risk.
Some studies have reported a higher incidence of MRSA infections in rural populations compared to urban areas. This could be attributed to the demanding work in rural areas, such as agriculture, inadequate personal hygiene practices, and the absence of surgical units in rural hospitals. Hospitals are one of reason for spreading of MRSA is Sewage pollution in environment. Factors such as the age range of individuals seeking medical attention, the presence of underlying diseases, the hospitalization duration, and adherence to hygiene and personal care principles in contact with patients may play a role in establishing this correlation.
The study focused on isolating MRSA from various samples in different hospital departments, with the ICU having the highest MRSA isolation rate at 24.34%. Concerning the unique conditions of the ICU and the patients admitted, it emphasizes the need for fundamental strategies and adherence to health care principles. In a study conducted in Mashhad (2016), out of a total of 7335 bacteria isolated from hospitalized patients at Imam Reza Hospital, the prevalence of 382 MRSA cases was reported (7.41%) (17). Studies by Madani in Saudi Arabia and Udo in Kuwait have reported that most MRSA isolates were obtained from blood and wound cultures from burn and ICU departments, with prevalence rates of 11% and 12%, respectively (18,19). The results of this study, when compared with similar studies, indicate that the prevalence of MRSA strains in ICU, NICU, and burn units is generally higher than in other departments. This could be due to the prolonged hospitalization of patients in these units, multiple concurrent illnesses, excessive use of antibiotics for infection treatment, aggressive therapeutic approaches, and the transfer of colonized patients from one hospital to another. These risk factors increase colonization, leading to the emergence of resistant strains. Therefore, increased surveillance and the implementation of effective infection control measures in these departments are necessary and crucial.
MRSA strains are currently a significant health concern. Given the widespread prevalence of MRSA, the treatment approach has shifted to vancomycin. Since the 1980s, vancomycin has become the drug of choice for treating severe MRSA infections in many healthcare facilities. However, the worrisome emergence of Staphylococcus aureus strains with reduced sensitivity to vancomycin and other glycopeptides has been noted (3). The results of antimicrobial tests in this study indicate multidrug resistance in these isolates. All MRSA isolates were resistant to cefoxitin and susceptible to linezolid. In the study by Rezazadeh, all MRSA strains were resistant to penicillin and susceptible to mupirocin (20). Asghari et al. reported the highest resistance to penicillin (96.6%) and erythromycin (45%) among MRSA isolates from clinical samples (21). Haddadi et al., in their 2016 study on clinical samples, showed that out of 250 clinical samples, 52 isolates of Staphylococcus aureus were identified, and antibiotic sensitivity results indicated that 68% (34 samples) were resistant to methicillin, with 24% (12 samples) resistant to ciprofloxacin (22). Rahimi reported a 30% prevalence of methicillin-resistant strains in three referral hospitals in Tehran (23). The highest antibiotic resistance in this study was reported against penicillin, clindamycin, tobramycin, and tetracycline, respectively.
The data suggest that the primary cause of the high prevalence of MRSA strains in hospitals and the reduced prevalence of susceptible strains may be due to the unnecessary prescription of various antibiotics for patients in hospitals or communities. Therefore, several specific genes in MRSA strains may adapt to hospital environments and continue to grow more than other strains developed in hospitals.
In conclusion, the emergence and prevalence of MRSA strains in hospital settings are influenced by various factors, including patient demographics, hospital unit conditions, and antibiotic prescription practices. The study highlights the need for enhanced surveillance, rigorous infection control measures, and a reassessment of antibiotic prescription practices to address the increasing resistance observed in MRSA strains.
In the present study, 5 (6.84%) of the isolated MRSA strains were found to carry the vancomycin resistance gene. A three-year study conducted in Iran on the prevalence of VRSA among Staphylococcus aureus isolates from 1393 to 1396 revealed that out of 1798 samples, four isolates (0.22%) were VRSA (24), and two isolates (0.11%) were VISA. In a surveillance study, all MRSA strains obtained were sensitive to vancomycin, with the highest resistance (4.84%) (25). observed against erythromycin. In a review study conducted by Nadri Nasab et al. until 2011, 29 cases of VRSA with the van gene were reported worldwide, including 12 cases from the United States, 16 cases from India, and one case from Iran (26). A study in Nigeria demonstrated that 277 out of 355 tested Staphylococcus aureus isolates (6.76%) were sensitive to vancomycin (27).
In Ethiopia, a study conducted in 2016 on 1360 samples revealed that out of 194 Staphylococcus aureus isolates, 34 (5.17%) were MRSA, and 10 (1.5%) showed resistance to vancomycin (28). A study in Shahid Beheshti Hospital in 2013, using the Agar method and PCR, identified five VISA isolates and one VRSA isolate among 63 clinical isolates with MIC ≥16μg/ml and the presence of the vanA gene (29).
Concerning the primary necessity to investigate vancomycin resistance through MIC, and using vancomycin disks in our study, it is possible that transitioning to the E-test strip could alter resistance results or reveal intermediate states, given the presence of the vanA gene in five MRSA isolates. Despite this, the study results indicate that the prevalence of susceptible strains or VISA with the vanA gene poses a significant risk for antibiotic resistance and the treatment of MRSA.
Various mechanisms contribute to antibiotic resistance in Staphylococcus aureus, with one important mechanism being the presence of efflux pumps in the bacteria. Over the past two decades, resistance of efflux pumps to fluoroquinolones such as ciprofloxacin, norfloxacin, and sparfloxacin has been described in clinical isolates of Staphylococcus aureus. Nor is a member of the efflux pump family, and the expression levels of nor efflux pump genes vary among different strains (30). In this study, 43 (90.58%) of MRSA showed resistance to ciprofloxacin. The frequency of the norA gene in ciprofloxacin-resistant samples was 13.58%, norB was 23.30%, and norC was 90.27%. Simultaneous presence of all three efflux pump genes or pairs of them was observed in some isolates.
In a study conducted in 1402, out of 100 clinical samples, 36 isolates of Staphylococcus aureus were recovered, with sensitivity tests indicating that 20 of them were resistant to ciprofloxacin. Fifteen isolates were resistant to norfloxacin, and one isolate was resistant to ofloxacin. Moreover, the norA, norB, and norC genes were found in 58%, 30%, and 27% of ciprofloxacin-resistant isolates, respectively (31). Other studies showed varying resistance frequencies, possibly due to different patient criteria in sample collection (32,33).
Another study by Afzal et al. showed that 56% of S. aureus isolated from eye infections were phenotypically resistant to ciprofloxacin. Also, 21.42% of resistant isolates have a high expansion in the norB gene (34). The results are slightly different from our research, which is probably due to the different criteria of patients in sample collection. In Hadadi's study, the frequency of norA and norB genes in ciprofloxacin resistant strains was 100% and 83%, respectively. Also, the efflux pump of all resistant strains was phenotypically active. Haddadi's study showed that among the strains with active pumps, almost half of them (48%) had higher expression. 57% showed increased expression in a single pump gene, usually norA, while the remaining 43% of isolates had increased expression in 2 or more output pumps, most of which were norB/C, and is associated with increased bacterial resistance to fluoroquinolones (22). Costa et al. confirmed the main role of the efflux pump as the first agent of antimicrobial resistance in Staphylococcus aureus (35). A plausible explanation for this could be that these isolates had enough pumps in their cell walls before the bacteria were exposed to the drug so that there was no need to increase the expression of the relevant genes. But overall, the results of this study with other studies show that the widespread use of fluoroquinolones in the treatment of this bacterial infection in the hospital is the most important factor in the development and spread of Staphylococcus aureus resistance to fluoroquinolones, and it has been suggested that the nor pump genes Efflux is one of the factors that cause resistance.
Conclusion
To sum up, control measures to prevent the spread of MRSA involve breaking the bacterial transmission chain (such as isolating clones or infected patients, hand washing, and patient screening) and efforts to reduce selective pressure related to MRSA by increased antibiotic use. Hospital hygiene guidelines are essential to prevent nosocomial infections, and health authorities should be informed about the potential consequences of Staphylococcus aureus infections inside and outside hospitals, reducing carriers. The rapid spread of resistant strains in hospitals is a known concern, and this study highlights the risk of such infections in hospital systems. Due to the clinical significance of these infections, it is imperative for healthcare professionals to be aware and plan to improve methods of identification and control of these infections in healthcare systems.
Acknowledgements
The authors would like to sincerely thank the laboratories staffs in hospitals especially to Mr.Alireza Mordadi to transferring samples to faculty of alliedof medical sciences.
Abbreviations
MRSA: Methicillin Resistant Staphylococcus Aureus
VRSA: Vancomycin-Resistant Staphylococcus Aureus
VISA: Vancomycin Intermediate Staphylococcus Aureus
MDR: Multi-drug Resistant
PCR: Polymerase Chain Reaction
MIC: Minimum inhibitory concentration
LZN: Linezolid
IPM: Imipenem
CIP: Ciproflaxin
AMK: Amikacin
FOX: Cefoxitin
CLI: Clindamycin
CTX: Cefotaxime
ERY: Erythromycin
CRO: ceftriaxone
CFZ:L Cefazolin
SXT: Trimethoprim / Sulfamethoxazole
VA: Vancomycin
Funding
No funding was received from profit and non-profit organization to do this article.
Authors' Contributions
Monireh Rahimkhani and Zahra Rajabi were contributed to the study design, data collection, analysis, and drafting of the manuscript. Saeed Alavi, shokoofeh Akbari and Manijeh Aliasghar were involved in practical part of research.
Conflicts of Interest
The authors declare that there is no conflict of interest associated with this article.
Orcid:
MonirehRahimkhani*: https://orcid.org/0000-0002-8663-7402
Zahra Rajabi: https://orcid.org/0000-0003-4970- 3068
Seyed saeed Alavi Fard: https://orcid.org/0009-0005-8819-7374
Shokoofeh Akbari: https://orcid.org/0009-0001-7561-3579
Manijeh Aliasghar: https://orcid.org/0009-0001-2403-9482
----------------------------------------------------------------------------------
How to cite this article: Monireh Rahimkhani*, Zahra Rajabi, Seyed Saeed Alavi fard, Shokoofeh Akbari, Manijeh Aliasghar, Investigation of efflux pump genes in staphylococcus aureus isolated from clinical samples. Journal of Medicinal and Pharmaceutical Chemistry Research, 2024, 6(5), 638-652. Link: https://jmpcr.samipubco.com/article_187679.html
----------------------------------------------------------------------------------
Copyright © 2024 by SPC (Sami Publishing Company) + is an open access article distributed under the Creative Commons Attribution License(CC BY) license (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
