Document Type : Original Research Article
Authors
- Fabricio Rogel Prado 1
- Riveliño Ramón Curay 1
- Jenny Martínez Moreira 1
- Joao Mazón Vélez 1
- Favian Bayas-Morejón 2
1 Universidad Estatal de Bolívar, Facultad de Ciencias Agropecuarias, Carrera de Medicina Veterinaria, CP: 020150, Guaranda, Ecuador
2 Department of Biotechnology, Universitat Politècnica de València, Valencia, Spain
Abstract
Bovine mastitis is of high significance in public health due to its potential as a source of infectious agents transferable between animals and humans. Extended-spectrum beta-lactamases (ESBLs) play a crucial role by hydrolyzing penicillins, cephalosporins (excluding cephamycins), posing a current public health problem. This study focused on detecting ESBLs in Escherichia coli and Klebsiella spp. isolated from bovine mastitis cases, using the Kirby-Bauer technique and phenotypic methods to assess antibiotic susceptibility, for which the data were collected in the microbiology area of the general laboratory of the State University of Bolivar for 12 months. Two E. coli strains were sensitive to all antibiotics, while two others showed resistance to the third-generation cephalosporins, with an increased inhibition zone in the presence of beta-lactamase inhibitors. The remaining two strains exhibited total resistance. In Klebsiella spp., all six strains were resistant to the third-generation cephalosporins, with an increased inhibition zone observed in one strain. 25% of the isolates were ESBL producers (33.33% in E. coli and 16.67% in Klebsiella spp.), with seven isolates carrying other beta-lactamases. In conclusion, bovine mastitis, as a source of infectious agents and the presence of beta-lactamases, stands out as a public health issue. The study highlights antimicrobial resistance, emphasizing the need for control and surveillance.
Graphical Abstract
Keywords
Main Subjects
Introduction
Within the components of the welfare of dairy cows, health, and the environment are pillars intimately connected to each other, with potential implications for the development of infectious diseases. Primarily, the most relevant pathologies in this productive stratum revolve around the cow's udder, where mastitis is the primary concern. Bacterial agents, as indicated by Bianchi et al. [1], play a major role, highlighting the importance of this condition.
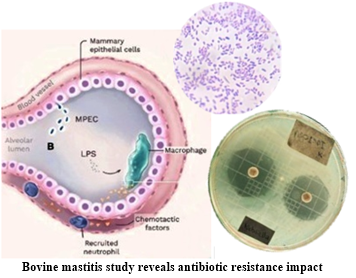
Bovine mastitis, an inflammation of the mammary glands in dairy cattle, is a multifaceted problem that impacts both the livestock industry and public health beyond its economic impact due to the reduced milk production and the associated costs of treatment, bovine mastitis has emerged as a serious challenge due to its connection to antibiotic resistance and its potential threat to human health [2,3]. Among the pathogens responsible for this disease, Escherichia coli and Klebsiella spp. occupy a prominent place [4]. Typically, Gram-negative environmental pathogens such as Escherichia coli and Klebsiella spp. are potential etiologies that cause significant acute and hyperacute mastitic conditions associated with environments featuring poor sanitary management. Each of these pathogens possesses adaptive characteristics and virulence factors, including the production of cytotoxic substances and enzymes that confer resistance to the animal's immune components and antibiotics [5].
Bovine mastitis poses a multifaceted challenge for both dairy producers and healthcare professionals. From the livestock industry perspective, the disease results in a significant decrease in milk production, translating into considerable economic losses [6].
From the public health perspective, bovine mastitis raises a series of additional concerns. The pathogens transmission from livestock to humans is a growing concern, as exposure to antibiotic-resistant infectious agents can have serious health implications. Antibiotic-resistant pathogens can complicate the treatment of infections in both animals and humans, underscoring the importance of understanding and addressing antibiotic resistance in the context of bovine mastitis [7,8].
The discovery of penicillins by Alexander Fleming in 1928 marked the beginning of antibiotic therapy to treat infections. Mastitis was not an exception to this therapeutic strategy, as antibiotics became the first choice for its treatment. However, over time, due to poor management of these medications, cure rates have declined. This is partly due to the ability of certain pathogens to develop resistance to antibiotics [9].
The ESBL role in antibiotic resistance
Antibiotic resistance is a global issue that affects the effectiveness of medical treatments in both humans and animals. Extended-Spectrum Beta-Lactamases (ESBLs) play a crucial role in this phenomenon, as they have the ability to hydrolyze the beta-lactam rings present in antibiotics, rendering them inactive and enabling pathogens to resist their action [10-12]. The ESBLs production by pathogens such as Escherichia coli and Klebsiella spp. is a particular concern, as these bacteria are responsible for various infections in both livestock and humans [13].
The presence of ESBL in strains of Escherichia coli and Klebsiella spp. in cases of bovine mastitis adds an additional layer of complexity to the problem. Resistance to third-generation cephalosporins, conferred by ESBL, limits treatment options and may lead to the failure of antibiotic therapies [14]. Furthermore, there is concern that these resistant pathogens may be a potential source of antibiotic resistance transmitted through the food chain and human exposure [15].
This study aims to characterize Escherichia coli and Klebsiella spp. susceptibility to antibiotics and identifies Extended-Spectrum Beta-Lactamases in strains from bovine mastitis cases. Using Kirby-Bauer and phenotypic methods, their response is assessed for common antibiotics. Understanding antibiotic resistance is crucial for developing effective strategies in preventing, treating, and controlling bovine mastitis.
Materials and methods
This study was carried out in the microbiology section of the General Laboratory within the Faculty of Agricultural Sciences, Natural Resources, and the Environment at the State University of Bolívar. Data collection spanned 12 months, involving isolates of Escherichia coli and Klebsiella spp. obtained from the department's microorganism bank.
Treatments and study factors
For the development of the present study, a Completely Randomized Design (CRD) was used according to the treatments shown in Table 1:
Revival of the bacterial isolates
Lab-cryopreserved isolates were revived following a specific procedure. Cryovials were allowed to equilibrate at room temperature for 30 to 60 minutes, followed by an incubation period of 30 to 60 minutes. Subsequently, a 10 µL aliquot of the revived bacteria was inoculated into MR-vP broth tubes, sealed with parafilm, and incubated at 37 °C for 24 hours.
Culture media such as MacConkey agar and EMB agar were used for initial plating. One hundred µL of the content from the incubated tube was streaked onto these media, which were then sealed and incubated at 37 °C for 24 to 48 hours.
This process of revival and plating on specific media is crucial for preparing the bacteria for subsequent study and analysis in the laboratory.
Biochemical identification
After obtaining purified isolates with uniform growth, the identification and confirmation of the bacteria were carried out using IMViC tests (Indole, Methyl Red, Voges-Proskauer, Citrate) and Triple Sugar Iron (TSI) profiling.
Antimicrobial activity
In the assessment of antimicrobial activity, Müeller-Hinton agar with a bacterial density of 1x10^8 CFU/mL (equivalent to a MacFarland scale of 0.5) was used. Colonies were spread across the entire surface of the medium using a sterile swab, followed by the application of antibiotic disks, and then incubated at 37 °C.
The antibiogram tests allowed the identification of extended-spectrum beta-lactamase production patterns, including the following tests: Susceptibility tests; Growth tests; Synergistic effect tests; and Zone of inhibition enlargement tests.
Antibiotic disks (Cefpodoxime 10 µg, Ceftazidime 30 µg, Aztreonam 30 µg, Cefotaxime 30 µg, and Ceftriaxone 30 µg) were used along with disks of Amoxicillin-clavulanic acid, Ceftazidime-clavulanic acid, and Cefotaxime-clavulanic acid to carry out the study.
According to CLSI-M1000- Ed32, [16], an increase of ≥ 5 mm in the diameter of the zone for any antimicrobial agent tested in combination with clavulanate compared to the zone diameter of the agent when tested alone is indicative of extended-spectrum beta-lactamase (ESBL) production. Tables 2 and 3 present the breakpoints for antimicrobial activity against the studied pathogens.
Estimation of prevalence
Prevalence of Escherichia coli and Klebsiella spp. producing ESBL in samples of mastitic bovine milk was estimated through the measurement of inhibition zones in the presence of Cefpodoxime, Ceftazidime, Aztreonam, Cefotaxime, and Ceftriaxone, as well as synergy tests with Amoxicillin and clavulanic acid. This approach allowed determining the relative frequency of bacteria showing resistance to third-generation cephalosporins in the context of bovine mastitis. The prevalence rate (PR) was calculated as the percentage of isolates producing ESBL among the total analyzed isolates using Equation (1).
Statistical analysis
The results obtained from the antimicrobial activity were tabulated and analyzed using the statistical software SAS version 9.4 to observe the effect of treatments on the experimental units (isolates).
Results and discussion
Effect of Beta-lactams against Escherichia coli
Based on the statistical analysis, in the antimicrobial activity assays using the Kirby-Bauer method, different measures of inhibition zones were observed. With a coefficient of variation of 5.41%, demonstrating the reliability and accuracy of data.
According to the 5% Duncan analysis of inhibition zones (Table 4), it was observed that Treatment 5, using 30 µg cefotaxime, showed the highest average of 26 mm. However, it is important to note that this treatment did not statistically differ from Treatment 6, which used ceftriaxone (mean of 25.33 mm), nor from Treatment 4, which employed 30 µg aztreonam (mean of 24.16 mm).
On the other hand, Treatment 3, with 30 µg ceftazidime, had a value of 22.33 mm, indicating statistical differences compared to those mentioned above. Treatments 2 and 1 showed the lowest average inhibition zones, with measurements of 17 mm and 16.50 mm, respectively, and were considered statistically equal.
In the study by Muleme [17], which examined Extended-Spectrum Beta-Lactamase (ESBL) producing strains of Escherichia coli isolated from humans, animals, and the environment, 9 strains from cows were found to exhibit resistance to third-generation cephalosporins. These strains showed significant differences in their resistance profiles, with measurements below the cutoff points established by the CLSI to be considered sensitive. These results are similar to those obtained in other strains, which also displayed variations in inhibition zones in antibiotic resistance profiles.
Study of the susceptibility of Escherichia coli against the drugs under study
In the susceptibility study of Escherichia coli isolates shown in Table 5, variations in the response to different antibiotics were observed. 83.33% of the strains were sensitive to amoxicillin plus clavulanic acid, according to the guidelines of the Eucast Disk Diffusion Method [18], and 33.34% according to document M100 of the CLSI-M1000- Ed32 [16]. However, 16.67% were considered resistant according to EUCAST and 66.68% had intermediate resistance according to CLSI.
Regarding the third generation cephalosporins, such as cefpodoxime, ceftazidime, cefotaxime, ceftriaxone, and aztreonam, it was observed that 33.34% of the isolates were susceptible, while 66.67% were resistant to each of these antibiotics. These results indicate that a significant proportion of the Escherichia coli strains studied showed resistance to the third-generation cephalosporins and aztreonam, underscoring the importance of addressing antibiotic resistance in this context.
In the study by Gelalcha & Kerro [13] about the prevalence of extended-spectrum beta-lactamase-producing Escherichia coli and antimicrobial resistance in dairy farms, it was found that of 14 Escherichia coli isolates obtained from milk from collection tanks, only 35.7% were resistant to cefotaxime (CTX). However, genomic analysis revealed the presence of genes that conferred resistance to cefotaxime and ceftiufur. These bacteria could be transmitted to humans through food.
In a study by Widodo et al. [19] on antimicrobial resistance and extended-spectrum beta-lactamase (ESBL) production in Escherichia coli on dairy farms in Indonesia, 124 isolates were identified, of which 87 were Escherichia coli isolated from cows' milk. Only 22.22% of these isolates (n=2) produced ESBL and showed total resistance to aztreonam and third-generation cephalosporins. These findings raise concerns about the threat they pose to public health.
When comparing the results obtained in the current study with the aforementioned results from the two previously cited investigations, it can be mentioned that there is significant variability in data from place to place. However, we agree that these isolates represent a potentially transmissible risk to public health, as therapeutic approaches in both human and veterinary medicine may be misguided and could potentially enhance the expression of virulence factors.
Effect of beta-lactams investigated against Klebsiella spp.
In the statistical analysis, different measures of inhibition zones were observed in the antimicrobial activity assays using the Kirby-Bauer method with a coefficient of variation of 10.95%, providing reliability and accuracy of data.
According to Duncan's analysis at 5% of the inhibition zones (Table 6), treatment 5, which used 30 µg cefotaxime, showed the highest average of 24.50 mm. It is important to note that this treatment did not differ statistically from treatment 4, which used aztreonam and had an average of 22.33 mm. Treatments 3 and 6, which used ceftazidime and ceftriaxone, respectively, showed statistical equality in their inhibition zone averages, being 19.67 mm for treatment 3 and 19 mm for treatment 6.
Finally, treatments 1 and 2, comprising amoxicillin plus clavulanic acid and cefpodoxime, respectively, exhibited statistical equality. Numerically, treatment 1 had the highest average with 16.33 mm, followed by treatment 2 with an average of 15.67 mm in the inhibition zone. These results highlight differences in the effectiveness of treatments in the formation of inhibition zones.
Demirci et al. [20] investigated genes related to the production of extended-spectrum beta-lactamases in Klebsiella spp. in raw milk from cows and isolated 4 strains of Klebsiella spp. They found that 50% of the isolates were resistant to ceftriaxone, and 1 isolate was resistant to ceftazidime and cefotaxime, with values of ≤27 mm in the diameter of the zone of inhibition. Compared to other research, this study showed a higher degree of resistance.
Study of the antimicrobial susceptibility of Klebsiella spp.
Different levels of susceptibility to antibiotics were observed in Klebsiella spp. isolates (Table 7). Regarding amoxicillin plus clavulanic acid, 100% of the strains were sensitive according to the EUCAST, but only 16.67% were sensitive according to the CLSI, with 83.33% of intermediate resistance. Regarding third generation cephalosporins, all strains were resistant to cefpodoxime, ceftazidime, cefotaxime, and aztreonam. However, against ceftriaxone, 16.67% were sensitive, and 83.33% were resistant. These results highlight the high resistance of Klebsiella spp strains to the third-generation cephalosporins and indicate the importance of addressing antibiotic resistance in this context. In the study by Borin Nobrega et al. [21] on the phenotypic and molecular characterization of extended-spectrum beta-lactamases produced by Klebsiella spp. in dairy cows, 28 strains of Klebsiella pneumoniae were isolated from 1547 milk samples, with an estimated prevalence of 0.60 cases per 100 cows. Of these strains, 2 isolates from mastitis samples and 1 from bulk tank milk tested positive for ESBL and exhibited resistance to third-generation cephalosporins. These findings are similar to those obtained in the present investigation, where resistance was also observed in all cephalosporins used.
In the research by Enferad & Mahdavi [22] on antibiotic resistance patterns and the frequency of genes associated with beta-lactamase production in Klebsiella pneumoniae isolated from raw milk samples in Iran, it was found that 5% of the studied population exhibited resistance to all tested antibiotics, including beta-lactams recommended by CLSI for characterizing extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae. Once again, these results are similar to those obtained in the present investigation, where total resistance to the beta-lactams used was observed, although only one isolate of Klebsiella spp. expressed sensitivity to the beta-lactamase inhibitor (clavulanic acid), a characteristic of ESBL-producing strains.
Characterization of the production of extended-spectrum beta-lactamases by Escherichia coli and Klebsiella spp.
As listed in Table 8, two strains of Escherichia coli and one isolate of Klebsiella spp. were identified as producers of extended-spectrum beta-lactamases (ESBL) due to their resistance to the third-generation cephalosporins, monobactams and penicillins, according to the phenotypic parameters proposed by CLSI-M1000- Ed32 [16]. The synergy test revealed an increase in the inhibition zone of ≥5 mm when ceftazidime (CAZ 30 µg) and cefotaxime (CTX 30 µg) were used in combination with clavulanic acid as a beta-lactamase inhibitor.
In addition, a plate growth test was carried out with medium containing 1 mg/L of ceftriaxone, which allowed us to observe that the strains of Escherichia coli and Klebsiella spp. identified as ESBL producers grew in this environment, demonstrating their capacity for resistance and contributing to their characterization. The strains that did not meet the parameters to be classified as extended-spectrum beta-lactamase (ESBL) producers showed no variations in the halo measurements in interaction with ceftazidime (CAZ 30 µg) and cefotaxime (CTX 30 µg), either alone or in combination with clavulanic acid. These strains were considered as producers of other types of beta-lactamases or as false positives for ESBL. The possibility of coexistence of ESBL and AmpC-type beta-lactamases was mentioned, which would result in resistance to all beta-lactams, as noted by Del Valle Martínez Rojas [14].
Among the Escherichia coli isolates, two strains were categorized as sensitive to the drugs used, according to the cut-off points for the interpretation of the phenotypic mechanisms of beta-lactamase production. These results allowed us to classify these strains as non-ESBL producers.
The study by El-Mohandes et al. [23] in mastitic cows on dairy farms in Egypt identified 80.48% of Escherichia coli isolates in cases of clinical mastitis, of which 38.2% were determined as producers of extended spectrum beta-lactamase (ESBL) by phenotypic diffusion tests. These findings are similar to those obtained in the present investigation, although the values may vary depending on the level of production and intensification.
Estimation of the prevalence rate of extended-spectrum beta-lactamase (ESBL)-producing isolates
The phenotypic characterization of extended-spectrum beta-lactamase (ESBL) producing strains revealed that, in the current investigation as shown in Table 9, 33.33% of Escherichia coli strains and 16.67% of Klebsiella spp. were classified as ESBL, which added up to a total prevalence of 25%.
Comparing these results with the study by Gelalcha & Kerro Dego [13], where they gathered data on extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae on dairy farms in the US, a significant variability in ESBL prevalence is observed. The reported data range from 20.5% on farms in Tennessee to 50% on farms in Washington for Escherichia coli ESBL. In addition, a prevalence of 2.8% is mentioned for Klebsiella pneumoniae ESBL on four farms in New York and 6.6% in positive samples from clinical mastitis on farms in California.
The increasing trend of extended-spectrum beta-lactamase (ESBL)-producing strains in dairy cattle identified in the cited study poses a significant risk to public health, as some of these bacteria are zoonotic and can be transmitted to humans in various ways. In addition, the antimicrobial resistance genes they carry can be transferred to other bacteria, increasing antibiotic resistance and potentially leading to severe therapeutic consequences. Anthropozoonotic transmission, involving farm workers and personnel, is an aspect that requires greater attention, especially in countries like Ecuador, where third-generation cephalosporins are used inhuman medicine. This raises concerns about the potential spread of resistant strains through the milk production chain.
Conclusion
In methodological terms, the research confirmed the identification of the 12 isolates under study, where 6 of them were classified as Escherichia coli and the remaining 6 as Klebsiella spp., ensuring the appropriate selection of strains for the study.
The results obtained through the analysis of pharmacological interactions in the antibiogram, using the CLSI (2023) and EUCAST (2023) breakpoints, revealed that 2 of the Escherichia coli isolates showed sensitivity to all evaluated drugs, while the remaining 4 isolates demonstrated resistance to the same drugs. In the case of Klebsiella spp. isolates, it was observed that all 6 isolates exhibited total resistance to all tested drugs.
Phenotypic characterization revealed that 3 isolates expressed the production of extended-spectrum beta-lactamases (ESBL), while the remaining 7 isolates produced other types of beta-lactamases. This observation suggests the coexistence of various types of beta-lactamases, which could complicate the precise detection of ESBL.
Regarding the prevalence of beta-lactamase production in the research, a total value of 25% was established. Of the total isolates, 33.33% (2/6) of Escherichia coli strains were categorized as ESBL producers, while 16.67% (1/6) of Klebsiella spp. strains manifested this resistance mechanism. These findings underscore the importance of addressing antibiotic resistance in the studied strains and the need for measures for their control and prevention.
Acknowledgements
We appreciate the Universidad Estatal de Bolívar, especially the general laboratory of the Facultad de Ciencias Agropecuarias, as well as the phytochemistry laboratory of the Vicerrectorado de Investigación y Vinculación for the support provided in the development of this research.
Funding
The research was funded by the authors and the Universidad Estatal de Bolívar.
Authors' Contributions
Fabricio Rogel Prado: Contributed to fieldwork and data collection; Riveliño Ramón Curay: Provided support in English language translation; Jenny Martinez-Moreira: Collaborated in data interpretation; Joao Mazón Vélez: Assisted in data collection, experimental part, and results discussion; Favian Bayas-Morejón: Contributed to the statistical interpretation of results and English language revisión.
Conflicts of interest
The authors declare that they have obtained consent from the livestock owners for the conducted analyses. Furthermore, they declare that thre is no conflicts of interest in this study.
Orcid:
Fabricio Rogel Prado: https://orcid.org/0009-0006-9085-6049
Riveliño Ramón Curay: https:/orcid.org/0000-0001-6284-4223
Joao Mazón Vélez: https:/orcid.org/0009-0006-4762-3282
Favian Bayas-Morejón*: https://orcid.org/0000-0003-2920-7155
-------------------------------------------------------------------------------
How to cite this article: Fabricio Rogel Prado, Riveliño Ramón Curay, Jenny Martinez-Moreira, Joao Mazón Vélez, Favian Bayas Morejón*, Antimicrobial activity and in vitro identification of extended-spectrum beta-lactamases (esbls) produced by escherichia coli and klebsiella spp. isolated from bovine mastitis cases. Journal of Medicinal and Pharmaceutical Chemistry Research, 2024, 6(6), 767-779. Link: https://jmpcr.samipubco.com/article_189490.html
-------------------------------------------------------------------------------
Copyright © 2024 by SPC (Sami Publishing Company) + is an open access article distributed under the Creative Commons Attribution License(CC BY) license (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

.png)
.png)

.png)
.png)
.png)
.png)
.png)
.png)
.png)
