Authors
1 Department of Medical Laboratory Sciences, Faculty of Science, Al-Balqa Applied University, Al-Salt 19117, Jordan
2 Department of Pharmacy, Faculty of Pharmacy, The University of Jordan, Amman, Jordan
3 Pharmacological and Diagnostic Research Center (PDRC), Faculty of Pharmacy, Al-Ahliyya Amman University, Amman 19328, Jordan
4 Department of Medical Laboratory Sciences, Faculty of Science, Mutah University, Mutah 61710, Jordan
Abstract
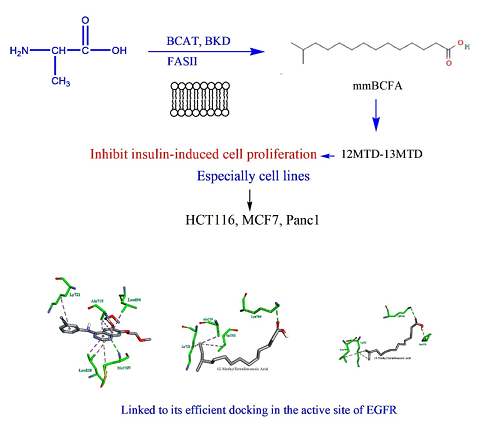
This study intends to investigate the potential use of mmBCFA, which includes 10 MTD, 12 MTD, 13 MTD, and 14 MTD, as a treatment option for cancer. This is due to the close correlation between metabolic dysregulation and the mmBCFA presence. We looked at how mmBCFA affected several cancer cell lines, such as those from the pancreas, colon, and breast. After determining the antiproliferative effects of these agents, we evaluated the expression levels of Bax, β-catenin, and cyclin D1. Furthermore, we performed Molecular Docking studies to look into how these agents interacted with the tyrosine kinase (EGFR) domain of the Epidermal Growth Factor Receptor. We discovered that mmBCFA efficiently inhibited the growth of multiple cancer cell lines, such as those from the colon, breast, and pancreas. Specifically, we found that when insulin was stimulated in HCT116, MCF7, and Panc1 cancer cells, 12MTD and 13MTD significantly reduced the expression levels of β-catenin and Cyclin D1. This was determined using selective qRT-PCR methods. In addition, our Molecular Docking analyses indicated that 12 MTD and 13 MTD interacted with specific residues in the EGFR domain. Based on our current findings, it appears that the mmBCFA supplementation could be considered as a novel and promising therapeutic option for treating cancer.
Graphical Abstract
Keywords
Introduction
Saturated fatty acids classified as BCFAs have one or more methyl branches along the carbon chain, with the main branch typically found closest to the end [1]. The branches can be identified as either iso- or anteiso-mono- methyl BCFA, with isopropyl or isobutyl groups respectively. Over 50 types of BCFA have been discovered in fats derived from ruminants [2].
You can find monomethyl branched chain fatty acids (mmBCFA) in a wide range of living things, including mammals and bacteria. Human skin produces the majority of BCFA, which are also present in internal tissues at low concentrations [3]. In addition, they can be found in breast milk and colostrum, where they account for about 1.5% of the total weight [4].
An essential function of the enzymes that break down branched chain amino acids (BCAA) is to control the synthesis of mmBCFA. A shared biosynthetic pathway converts the BCAAs into short chain branched acyl moieties. Fatty acid synthase (FAS) then uses these acyl moieties to carry out de novo fatty acid biosynthesis, producing mmBCFA that are subsequently integrated into unique lipid droplets. According to the recent research, the main process influencing the adipocytes' synthesis of mmBCFA is cytosolic de novo lipogenesis through FAS [5].
Human plasma contains monomethyl branched chain fatty acids (mmBCFA) at levels in the micromolar range. According to the recent research, the mmBCFA levels in plasma increase by 2-3 times during prolonged fasting, but they decrease in diet-induced obesity [6].
Researchers found that patients who are obese and insulin-resistant have varying amounts of mmBCFA. The results of the study showed that skeletal muscle insulin sensitivity and mmBCFA content were positively correlated, and that weight loss altered the mmBCFA content of adipose tissue. This suggests that mmBCFA may link the BCAA metabolism in fat tissue to insulin resistance associated with obesity in skeletal muscles [5].
Several studies have shown that mmBCFA has antitumor effects. Of these, the iso-C15 branched saturated fatty acid 13-Methyltetradecanoic acid (13-MTD) stands out for its capacity to damage mitochondria and trigger apoptosis in cancer cells. It has been demonstrated in earlier studies to suppress AKT phosphorylation, which is followed by caspase activation, to cause apoptosis and inhibit the growth of T-cell non-Hodgkin lymphoma (Jurkat, Hut78, and EL4 cell lines) [7]. Our goal in this work is to investigate the antiproliferative effects of particular mmBCFA on various cancer cell types and to suggest a possible molecular mechanism for this action.
In previous study, Moath Alqaraleh et al. (2019) proposed that both phytochemicals and mmBCFA could be utilized to treat diabetes and that co-cultivating the phytochemical compounds and mmBCFA could be a distinct therapeutic approach to treat diabetes and obesity-related colon cancer [8].
Hypothesis
Concerning the obese patients with lower concentrations of mmBCFAs than healthy individuals and since an inverse correlation was observed between the concentrations of mmBCFAs band serum concentrations of insulin, we hypothesize that mmBCFAs may affect the expression of genes related to FAs synthesis and inflammation. Our goal was to assess whether mmBCFAs may exert potential beneficial effects connected with these processes.
Materials and methods
Cancer culture
MCF7 (breast, mammary gland; derived from pleural effusion, a metastatic site) is one of the human cell lines used to treat breast cancer. ATCC HTB-22 and T47D (derived from the pleural effusion metastatic site; mammary gland, HTB-133), pancreatic cancer cell line Panc1 (ATCC CRL1469), and human colorectal cancer cell lines, including HT-29 (ATCC HTB-38), HCT116 (ATCC CCL-247), SW620 (ATCC CCL-227), SW480 (ATCC CCL-228), and CACO2 (ATCC HTB-37) were cultured in DMEM supplemented with 10% FBS, HEPES Buffer (10 mM), L-glutamine (100 μg/mL), gentamicin (50 µg/mL), penicillin (100 µg/mL), and streptomycin (100 mg/mL).
Anti-proliferative assay
The viability assay
The SRB colorimetric assay for cytotoxicity screening was used to determine cytotoxicity measurements and the mechanism of reduction of cell viability; this assay was previously described in [9]. HT29, SW480, SW620, HCT116, CACO2, MCF7, T47D, and Panc1 were among the cancer cell lines used in the experiment. The cells were seeded in 96-well plates with 5000 cells per well and cultured for a full day. The cells were cultured in a medium containing mmBCFA, such as 10 MTD, 12 MTD, 13 MTD, or 14 MTD, at concentrations ranging from 20-400 µM, either with or without insulin, following a 48-hour serum starvation period [10]. The SRB assay was carried out in accordance with Vichai et al. (2006) [11] after 72 hours.
The primary cell culture of human periodontal ligament fibroblasts (PDL) was utilized to get the lowest antiproliferative IC50 value to confirm selective cytotoxicity. Cisplatin (0.1-200 μg/mL), a well-known and potent anti-cancer drug, was utilized as a reference agent to compare the outcomes. Every experiment was carried out in triplicate, and the mean values ± SD (n=3) were used to report the IC50 antiproliferative activities.
Gene expression analysis
To assess changes in gene expression levels of Bax, Cyclin D1, and β-catenin, HC116, MCF7, and Panc1 cell lines were cultured in 6-well plates at a density of 1 × 106 cells per well, with or without insulin (250 nM). After a 24-hour incubation period, the cells were treated with branched chain fatty acids at a predetermined concentration and incubated for an additional 24 hours. The fold change in gene expression was then measured [12].
Extracting and analyzing ribonucleic acid (RNA)
Total RNA for this investigation was extracted using the RNeasy Mini kit (QIAGEN, USA). Cell pellets were initial defrosted and suspended in a 2-mercaptoethanol-containing lysis solution. Following the addition of ethanol, the lysate was put on an RNeasy Mini spin column to remove cellular debris and trap all of the RNA inside the binding column. After three rounds of washing, RNase-free water was used to elute the RNA. The concentration and purity of the extracted RNA were measured using the NanoDrop ND-1000 spectrophotometer, with the majority of the samples exhibiting a ratio (A260/A280) of 1.8-2.1.
Complementary deoxyribonucleic acid (cDNA) synthesis
The study utilized a reverse transcription system from Applied Biosystem, USA, to create cDNA. First, a microcentrifuge tube containing 2 μg total RNA and 1 μL oligodeoxythimidine primer was incubated and centrifuged. Next, a 20 μl reaction solution was created with several reagents, including AMV-RT, and thermal conditions were set for cDNA synthesis. The resulting cDNA was analyzed for concentration and purity using a NanoDrop ND-1000 spectrophotometer, with most samples having the ratio (A260/A280) of 1.6-1.8. The cDNA samples were then stored at -80 °C for additional examination.
Relative quantitative RT-PCR analysis
Using a fast SYBR green kappa master mix (Biosystem, USA), the relative-quantitative mRNA expression levels of particular genes were determined. A small amount (1-2 μL) of cDNA template, 1x concentrated KAPA SYBR green fast mastered mix, a forward primer and a reverse primer (each at a concentration of 200 nM), and a final volume of 20 μL were all included in the PCR reaction mixture. Table 1 lists the primers used in the reaction. The melting point was between 70 °C and 95 °C. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was employed as an internal reference gene to normalize the expression level of the examined genes. Three duplicates of the experiment were carried out.
Molecular docking
We utilized a range of specialized software programs and computational tools to conduct molecular docking simulations on several compounds, including a branched fatty acid, with the EGFR domain of the epidermal growth factor receptor, also known as the receptor tyrosine kinase. They started by obtaining the EGFR crystal structure from the Protein Data Bank's database [17]. After that, they used the ACD/ChemSketch software to create the compounds' chemical configurations. The branched fatty acid was saved as a mol file, and then converted into a pdb file using Avogadro software [18] before using AutoDockTools to prepare the ligand files in pdb format. Using AutoDock 4.2, the docking simulations were carried out. Kollman and Gasteiger charges were added to plant compounds and proteins, respectively, with grid maps created using AutoGrid 4 to map specific regions of the protein structure. The authors further applied the Lamarckian genetic algorithm (LGA) to optimize and minimize energy during the docking simulation.
To validate their docking parameters, they used the co-crystallized ligand (Structure Code: 1M17) in a molecular docking modeling. Totally, we employed a multi-step approach, leveraging various computational tools and techniques to perform a comprehensive analysis of the molecular docking of compounds with the EGFR domain [16].
Statistical analysis
The results were presented using the means and standard deviations from three to four separate experiments. Dunnett's post hoc test and GraphPad Prism ANOVA were utilized to assess statistical differences between the treatment and control groups. Statistical significance was defined as a p-value of less than 0.05, and high significance as a p-value of less than 0.001.
Results
mmBCFA modulates colorectal, cancer, and fibroblast cell line proliferation
When subjected to 250 nM insulin, unbranched PMA has been shown to lose the capacity to prevent the proliferation of cells in each kind of colorectal carcinoma, as presented in Table 2. However, 12MTD, 13MTD, 14MTD, 10MTD, and HT29 showed significant anti-proliferative effects against HCT116, SW620, Caco2, and SW480 over a 72-hour incubation period. The mmBCFA further demonstrated sustained anti-proliferative effects against HCT116, SW620, and SW480, but not against Caco2 when co-treated with insulin. Although the effective mmBCFA exhibited increased IC50 values when treated with insulin, they did not exhibit selective cytotoxicity in fibroblasts, as summarized in Table 2.
mmBCFAs' modulation of breast and pancreatic cancer cell lines' proliferation
The impact of cisplatin on the cytotoxicity of MCF7, T47D, and Panc1 cancer cells is indicated in Table 3. The antiproliferative impact of unbranched PMA on MCF7, T47D, and Panc1 was eliminated by insulin. Furthermore, the cytotoxicities of 12MTD, 13MTD, 14MTD, and 10MTD on MCF7, T47D, and Panc1 during 72-hour incubations were diminished when co-treated with insulin (Table 3). Compared to cisplatin, none of the mmBCFA exhibited the same level of potency.
Gene expression analysis for different cancer cell lines under the effect of mmBCFA
The selective effects of mmBCFA on beta catenin, Bax, and cyclin D expression in HCT116, MCF7, and Panc1 cell lines
Insulin was observed to upregulate β-catenin expression in MCF7, Panc1, and HCT116 cell lines. Nevertheless, as shown in Figures 1 and 2, the co-administration of 12MTD and 13MTD significantly reduced the amount of insulin-induced β-catenin expression. Furthermore, Figure 2 demonstrates that, although insulin decreased the expression level of the antiapoptotic protein BAX in HCT116, MCF7, and Panc1, the IC50 values of 12MTD and 13MTD significantly increased the BAX expression in MCF7 and Panc1, but not in HCT116. As demonstrated in Figures 3, insulin also increased the expression of cyclin D1 in HCT116, MCF7, and Panc1. However, when 12MTD and 13MTD were administered in conjunction with insulin, the expression of cyclin D1 was significantly downregulated in these cancerous cell lines.
Molecular docking against EGFR
The 1M17 crystal structure successfully accommodated the co-crystallized ligand (AQ4) with an RMSD of approximately 1.37 Å (less than 2.0 Å), as illustrated in Figure 4. Based on the research [19], molecular docking simulations with an RMSD value of less than 2.0 Å are considered efficient. To dock other compounds in the active site, similar parameters were employed. Figure 5 indicates that Met769 had hydrogen bond interactions with the AQ4 inhibitor, while hydrophobic and VDW interactions involved other amino acids such as Ly721, Ala719, Leu694, and Leu820 [19].
The docking of the prepared compounds against 1M17 was successful, and Table 4 illustrates the best binding energy results. The energy analysis revealed that compounds 12MTD and 13MTD interacted with the co-crystal structure (AQ4) via similar residues, as demonstrated in Figure 6.
Discussion
It has been established that insulin may play a role in promoting tumor growth in test animals. According to a number of observational studies, people with type 2 diabetes who receive insulin treatment have a higher incidence of cancer mortality [21-26], fueling concerns that insulin may play a role in the development of cancers or may be associated with increased mortality [27]. We carried out a study to look into this matter. We looked at how mmBCFA affected the growth of different cancer cell lines as well as fibroblast cell lines when insulin was administered chronically at low levels. Our results were unexpected because they demonstrated that mmBCFA could directly prevent HCT116, MCF7, and Panc1 cells from proliferating in response to insulin. We suggest that because mmBCFA inhibits insulin's anti-apoptotic effect, it increases apoptosis, which in turn inhibits the growth of colorectal, breast, and pancreatic cancers.
Previous research by Serra et al. (2008) and Mi et al. (2009) has shown that insulin is known to activate downstream signaling pathways, such as PI3K/Akt and MAPK, which are essential for controlling the cell cycle and preventing apoptosis [28,29]. We examined the expression of beta catenin, Bax, and Cyclin D genes using Real-time PCR during chronic insulin treatment in order to better understand the mechanisms underlying the suppressive effect of mmBCFA on insulin-induced cancer cell proliferation. According to our in silico study, mmBCFA's effective docking in the EGFR active site under the influence of insulin may be responsible for its suppressive effect on cancer cell proliferation, which includes the overexpression of the Bax gene and the downregulation of beta catenin and Cyclin D gene.
The PI3K/Akt and MAPK pathways, which control the cell cycle and prevent apoptosis, are activated by insulin [28,29]. In an effort to more successfully inhibit tumor growth, researchers are now investigating insulin resistance and IGF/IGF-IR axis inhibition as a possible strategy. This involves activating mTORC1/S6K1 to induce a negative feedback loop and inhibiting mTORC2 kinase activity toward Akt [28,29]. By blocking GSK via the Akt and MAPK pathways, IGF-1 and insulin can activate beta catenin. This stops proteasomes from breaking down beta catenin and enables its translocation from the cell membrane to the cytoplasm [30]. As a result, beta catenin builds up in the cytoplasm, where it is transferred to the nucleus to regulate the expression of target genes, including those involved in the progression of the cell cycle like Cyclin D [31,32]. Furthermore, proapoptotic genes like Bax, which cause mitochondrial membrane permeability, are expressed more frequently when beta catenin is present, and this is known to contribute to apoptosis [33].
Fascinatingly, it has been discovered that mmBCFA directly inhibit the growth of insulin-induced cancer cells in cell lines, including colorectal, breast, and pancreatic cancer cells, by promoting apoptosis and counteracting insulin's anti-apoptotic effect. This is most likely caused by GSK inhibition, which stabilizes and accumulates beta catenin in the cytoplasm, causing cell growth to be inhibited and apoptosis to be induced [34-36]. In addition, it has been demonstrated that mmBCFA inhibits the over-activation of PI3K/Akt induced by insulin and has growth-inhibitory effects by causing tumor cells to undergo apoptosis [37].
In this study, we have demonstrated that mmBCFA exhibits cytotoxic effects on various cancer cell lines, potentially achieved through the antagonism of insulin pathways such as the MAPK pathway and PI3K/Akt pathway. This observation leads us to propose that interference with these insulin pathways may occur through the inhibition of EGFR, thereby inducing an antiproliferative effect in cancer cells [38-40]. Activation of EGFR is known to trigger four major downstream signaling pathways, including MAPK/ERK, PLCγ/PKC, JAK/STAT, and PI3K/AKT, which influence transcriptional regulation and cell cycle progression, ultimately contributing to cancer and inflammatory diseases.
Hence, we hypothesize that the antiproliferative action of mmBCFA against cancer cell lines may stem from their ability to bind to the extracellular domain of the EGFR, competing with growth factors for receptor binding (Figure 6a and 6b). Consequently, through receptor blockade, treatment with mmBCFA is likely to impede tumor cell proliferation [40].
Conclusion
Our findings suggest that mmBCFA may counteract the anti-apoptotic effects of insulin, hence impeding insulin-induced cell proliferation in different cancer cell lines. This obstacle may arise from decreased expression of β-catenin and cyclin D1, along with elevated expression of BAX. Moreover, molecular docking studies suggest that mmBCFA's anti-proliferative effects may be related to their interaction with EGFR's active site, which sheds light on the efficacy of these agents. Furthermore, our results highlight the potential therapeutic benefit of mmBCFA in the treatment of pancreatic, breast, and colorectal malignancies, especially those involving insulin-related pathways. Therefore, examining the synergistic effects of mmBCFA with well-established cancer therapies such as chemotherapy or targeted medicines may enhance treatment results and tackle resistance mechanisms.
Acknowledgements
The authors would like to thank everyone helped in this valuable study.
Funding
This study did not receive any funding.
Authors' Contributions
M.A., V.K., K.K. designed and performed the experiments, derived the models and analysed the data. B.A. helped carry out the docking. M.A. and V.K. wrote the manuscript in consultation with K.K. and F.M.
Conflict of Interest
The authors declare that they have no competing financial interests or personal relationships that could have appeared to influence the work reported in this article.
Orcid:
Moath Alqaraleh: https://orcid.org/0000-0003-2072-2441
Violet Kasabri: https://orcid.org/0000-0003-1927-0193
Frezah Muhana: https://orcid.org/0000-0002-5318-0514
Belal Omar Al-Najjar: https://orcid.org/0000-0001-6811-1792
Khaled M. Khleifat: https://orcid.org/0000-0002-0216-3175
---------------------------------------------------------------------------------
How to cite this article: Moath Alqaraleh, Violet Kasabri, Frezah Muhana, Belal Omar Al-Najjar, Khaled M. Khleifat, Evaluation of the antiproliferative activity and molecular docking of selected branched fatty acids. Journal of Medicinal and Pharmaceutical Chemistry Research, 2024, 6(9), 1340-1353. Link: https://jmpcr.samipubco.com/article_193795.html
---------------------------------------------------------------------------------
Copyright © 2024 by SPC (Sami Publishing Company) + is an open access article distributed under the Creative Commons Attribution License(CC BY) license (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
