Document Type : Original Research Article
Authors
1 Doctoral Program of Medical Science, Faculty of Medicine, Airlangga University, Surabaya, 60132, Indonesia
2 Department of Anatomy, Histology and Pharmacology, Faculty of Medicine, Airlangga University, Surabaya, 60132, Indonesia
3 Faculty of Veterinary Medicine, Airlangga University, Surabaya, 60132, Indonesia
4 Department of Toxicology, Advanced Medical and Dental Institute, Universiti Sains Malaysia, Bertam, 13200, Penang
Abstract
Inflammation is often the key element that results in dysregulation of one or more of the biochemical pathways responsible for the pathological development of disease. Uncontrolled acute inflammation can become chronic, contributing to various chronic inflammatory diseases such as cardiovascular disease, diabetes, arthritis, and cancer. Primary stimulation is generally elicited by proinflammatory cytokines such as interleukin 1β (IL-1β), interleukin 6 (IL-6), and tumor necrosis factor α (TNF-α). Petiveria alliacea L. has been reported pharmacologically to have anti-microbial, anti-cancer, immunomodulatory, analgesic, and anti-inflammatory activities. The purpose of this study was to determine the Petiveria alliacea potential as an anti-inflammatory by inhibiting proinflammatory cytokine target proteins (IL1R and TNFAR) in silico. The test compounds are selected compounds obtained from the UPLC-QToF-MS/MS results of Petiveria alliacea leaf extract and the reference results from Google, which are compounds that have been widely studied. Prior to docking, the downloaded compounds were prepared using PyRx 0.8, and then docked with both receptors using AutoDock Vina, and visualization of the docking results was carried out using Biovia Discovery Studio 2019. The docking results showed that the Myricitrin compound had a lower binding affinity value than the IL1R receptor inhibitor, which indicated that it had better activity compared to the inhibitor as well as the isoarborinol acetate compound against TNFAR. Therefore, the conclusion of this study is that the 70% ethanol extract of Petiveria alliacea leaves has anti-inflammatory activity by inhibiting pro-inflammatory cytokines (IL1R and TNFAR).
Graphical Abstract
Keywords
Main Subjects
Introduction
Inflammation is a pathogenic event arising from the immune system's activation in response to various stimuli. Often, inflammation plays a significant role when one or more biochemical pathways responsible for the development of inflammation-related disorders become dysregulated [1,2]. Many chronic diseases, including cancer, diabetes, rheumatoid arthritis, cardiovascular ailments, intestinal disorders, and other forms of arthritis, share inflammation as a common underlying cause [3,4].
The body creates acute inflammation as a sort of short-term inflammation to address damage, illness, and infection [2]. Nevertheless, if acute inflammation is not addressed, it can progress to chronic inflammation, contributing to many chronic inflammatory diseases [4,5]. The existence of monocytes, lymphocytes, and macrophages, as well as the connective tissue and growth of blood vessels, are the primary features of chronic inflammation. An organism's inflammatory response might eventually start to harm healthy cells, tissues, and organs while living with chronic inflammation. Internal scarring, tissue death, and DNA damage are all its possible long-term effects [2].
Intracellular signaling pathways are activated by inflammatory stimuli, and this in turn activates the synthesis of inflammatory mediators. Primary inflammatory triggers like microbial products and cytokines, such as interleukin 6 (IL-6), interleukin 1β (IL-1β), and tumor necrosis factor α (TNF-α), drive inflammation through their interactions with receptors like TLRs, IL-6R, IL-1R, and TNFR. These interactions activate key cellular signaling routes, including the Janus kinase (JAK)/signal transducer and activator transcription (STAT) pathways, nuclear factor kappa B (NF-κB), and mitogen-activated protein kinase (MAPK) [4,6].
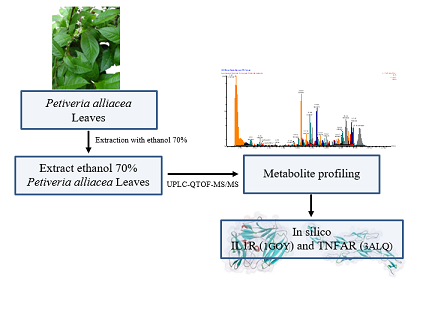
Guinea chicken grass, sometimes referred to as Petiveria alliacea L. (Phytolaccaceae), is a tropical medicinal plant. It is a perennial plant that grows wild in Central America, South America, and Africa [7]. P. alliacea is employed ethnopharmacologically to treat rheumatism, diabetes, and inflammation. It can also be used as a depurative, anesthetic, sedative, antispasmodic, antihelminthic, antispasmodic, and antinociceptive agent. P. alliacea has been shown to possess pharmacological properties that include anti-microbial, anti-cancer, immunomodulatory, analgesic, and anti-inflammatory effects [8-10]. This study aims to determine the metabolite profiling of P. alliacea extracts as well as their potential as anti-inflammatory agents by in silico analysis of these compounds with pro-inflammatory cytokine target proteins, such as IL1R and TNFAR.
Materials and methods
Extraction of petiveria alliacea leaves
Petiveria alliacea leaves were obtained and identified at the Unit Pelaksana Teknis (UPT) Materia Medika, Batu, East Java, Indonesia, with the identification letter of 074/349/102.7-A/2021, and then the leaves of Petiveria alliacea were extracted by maceration with 70% ethanol solvent in a ratio of 1:10, for 3x24 hours with occasional stirring.
Metabolite profiling using UPLC-QToF-MS/MS
Metabolite profiling was conducted at the Forensic Laboratory Center for the Indonesian National Police Criminal Investigation Agency using the UPLC-QToF-MS/MS system. The extract was prepared using the solid-phase extraction (SPE) method. Subsequently, 5 µl of each extract was introduced into the MS Xevo G2-S QToF detector, part of the ACQUITY UPLC® H-Class System (both by Waters, USA). Sample separation occurred on an ACQUITY BEH C18 column (1.7 µm 2.1 × 50 mm) with a flow rate of 0.2 ml/min, utilizing acetonitrile + 0.05% formic acid and water + 0.05% formic acid as the mobile phases. The UPLC-QToF-MS/MS analysis results were processed using the MassLynx 4.1 software, which generated chromatogram data and m/z spectra for each peak. Chemical identifications were further validated using online resources: MassBank (https://massbank.eu/MassBank) and ChemSpider (https://www.chemspider.com) [11].
Ligand preparation
The test compounds are selected compounds obtained from the UPLC-QToF-MS/MS results of Petiveria alliacea leaves extract and the reference results from Google, which are compounds that have been widely studied [10,12-13]. The 3D structures of the compounds in Table 1 were obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/).Compounds were prepared by optimizing their conformation using the Open Babel 2.3.1 plug-in integrated in the PyRx 0.8 software. The optimum conformation will make the ligand structure flexible [14,15]. After preparation, the compounds are stored in the Protein Data Bank (PDB) format.
Preparation of Inhibitor Compounds for Control
Inhibitor compounds are compounds that have the ability to inhibit a protein. The list of inhibitors used is indicated in Table 2. The inhibitor compounds were prepared in the same way as the active compounds.
Protein Preparation
When using the ID listed in Table 2, it is possible to download the 3D structures of the IL1R and TNFAR proteins from the RCSB PDB database (https://www.rcsb.org/) [16]. Using the Biovia Discovery Studio 2019 program, contaminating compounds were eliminated to prepare these proteins [17]. This is necessary because the contaminant molecules will hinder the protein binding process with the test ligand.
Molecular docking
Molecular docking was carried out using AutoDock Vina, which was integrated into the PyRx 0.8 software [18]. The selected docking result is the one with the lowest binding affinity value in each mode. The binding affinity value reflects the strength of biomolecular interaction between the ligand and the receptor, the smaller it is, the more stable it is and indicates the stronger affinity of the ligand for the receptor [19].
Chemical interactions
Visualization of the docking results was carried out using the Biovia Discovery Studio 2019 software [20,21]. The chemical interactions shown are hydrogen bonds, hydrophobic interactions, electrostatic interactions, and unfavorable interactions. Interactions that produce many hydrogen bonds are considered stable [11]. This is because hydrogen bonds are the strongest of all types of bonds in molecule docking [22].
Structural visualization
Visualization was performed using Discovery Studio 2019 software. Ligands and proteins are displayed in 3D structures to provide an overview of the ligands and proteins before they are interacted with [23,24]. The 3D visualization of protein-ligand interactions was carried out using whole visualization, and then focusing on the ligand-binding side of the protein using surface structures. Experiment should start as a continuation to introduction on the same page. All important materials used along with their source shall be mentioned. The main methods used shall be briefly described, with references. New methods or substantially modified methods may be described in sufficient detail. The statistical method and the chosen significance level shall be clearly stated.
Results and discussion
Metabolite profiling
Metabolite profiling was conducted to determine the composition of compounds in the Petiveria alliacea extract [25]. The extract was prepared using the Solid Phase Extraction (SPE) method before being profiled using a UPLC-QToF-MS/MS device. The advantage of using SPE for sample preparation is its ability to filter out impurities, enhancing the sensitivity of the spectral readings [26].
To avoid bias when identifying the sample, the total ion chromatogram (TIC) analysis of the blank was performed before the TIC of the chemicals in the sample. MassLynx 4.1 software was used to analyze the mass spectrum of each TIC peak, and the results were then verified against the ChemSpider and MassBank web databases. Figure 1 dispalys the total ion chromatogram (TIC) of the results of the metabolite profiling of Petiveria alliacea extract using the UPLC-QToF-MS/MS instrument. Table 1 presents the % area, compound name, retention time (RT), m/z, molecular formula, and its activities as determined by literature studies. Metabolite profiling with UPLC-QToF-MS/MS showed that an extract of Petiveria alliacea had a total of 70 compounds, 58 of which were known and 12 of which were unknown. Not all peaks in TIC could be recognized during the metabolite profiling method based on the total number of chemicals measured. The extract from Petiveria alliacea contains unidentified chemicals, which is a sign of this. Unknown compounds are substances that cannot be identified in the database, they can be impurities or degradants that are still detectable by instruments or new substances that have not yet been included in the database, particularly if they are present in large amounts [27].
Based on the analysis of these metabolites, there are a number of dominant or main compounds. These are compounds with more of them than other compounds in the sample, which is shown by the percent area. The major compounds in Petiveria alliacea extract are D-(-)-Morphine with an area percentage of 10.6739%; N2-(4-{[(2,4-Diamino-6pteridinyl)methyl](methyl)amino}benzoyl)-N-(1-methoxy-1-oxo-2-tetradecanyl)-L-glutamine with an area percent as much as 5.0316%; and Octadecatetraenoic acid with an area percent of 3.8440%. In addition, there are also several compounds that have activity as antioxidants and anti-inflammatories, such as Isoarborinol with an area percent of 3.5927%, Myricetin with an area percent of 0.4590%, Quercetin with an area percent of 0.4590%, and so on [28-33]. This shows that Petiveria alliacea extract has the potential to have activity as an antioxidant and anti-inflammatory.
Protein Structure
The list of proteins used in this study along with their PDB ID is inidicated in Table 2. The control compounds used in this analysis are also listed in Table 2. The control compounds are inhibitory compounds for each protein that have been discovered by previous researchers. The 3D structure of the protein is displayed in a ribbon style with secondary structure staining. The red color represents the helix structure, the light blue color represents the beta-sheet structure, the white color represents the loop structure, and the green color represents the coil structure (Figure 2).
Molecular docking
The type of chemical bond produced and the value of binding affinity have a strong correlation with docking results. The amount of power needed to bind a protein to its ligand is called binding affinity. The easier it is for the ligand to bind to the protein and the more potential it has to affect the protein, the lower the binding affinity value [34]. The results of the protein-compound docking yielded binding affinity values, as provided in Table 3. From these results, it can be seen that the IL1R-Myricitrin complex and TNFAR-Isoarborinol acetate have the most negative binding affinity values.
Docking results between the IL1R protein and the test compounds showed the test compound with the most negative binding affinity value (Figure 3). Myricitrin interacts at the same amino acid residue as the inhibitor, namely at Asn10 (Table 4). The docking results between TNFAR and the test compounds show that all the test compounds bind on the same side as the inhibitor (Figure 4). The same binding position as the inhibitor indicates that the test compound has similar activity to the Inhibitor, namely inhibiting TNFAR protein activity (Table 4) [35]. Isoarborinol acetate interacts with TNFAR by forming a hydrogen bond in Asn149. The residue is also the position of the inhibitor interaction on TNFAR.
Some of the classic cytokines that trigger the inflammatory response in general disease are IL-6, IL-1, TNF-α, and the IFN family, these have also been described in the pathology of diabetes, which provides clinical benefit by blocking these cytokines [36].
IL-1 is an inflammatory cytokine with numerous physiological and pathological roles that are crucial for preserving the balance between health and disease. Interleukin-1 has evolved, and mounting evidence emphasizes its significance in tying innate immunity to a wide range of disorders beyond inflammatory ones [37]. The IL-1 protein family encompasses several members: IL-1Ra, IL-18, IL-33, IL-36Ra, IL-36γ, IL-36β, IL-36α, IL-37, IL-38, IL-1α, and IL-1β [38,39]. Among these, the ones that act as receptor agonists are IL-18, IL-33, IL-36, IL-1α, and IL-1β. Conversely, IL-36Ra, IL-38, and IL-1Ra serve as receptor antagonists. Notably, IL-37 stands out as the sole anti-inflammatory cytokine [37].
By regulating a number of innate immune functions, IL-1 is the primary regulator of inflammation [37,39]. From a historical perspective, IL-1 has various biological effects, such as serving as a leukocyte pyrogen, a fever mediator, an endogenous leukocyte mediator, and an inducer of many acute phase response components and lymphocyte activating factor (LAF) [37]. In addition to IL-1, TNF-α plays a pivotal role in the inflammatory process. It has been observed that TNF-α boosts the expression of MHC I molecules, thereby hastening cell apoptosis [36]. In the diabetes context, IL-1 induces spurs local inflammation and b-cell apoptosis. On the other hand, TNF-α promotes speeds up b-cell apoptosis, dendritic cell maturation, and activates antigen-specific T cells [36]. Therefore, inhibition of inflammatory cytokines is very important in the pathology of disease development.
Conclusion
In silico analysis, 70% ethanol extract of Petiveria alliacea leaves has anti-inflammatory activity by inhibiting pro-inflammatory cytokines (IL1R and TNFAR). Based on the binding affinity value, the compounds with the most potential as IL1R and TNFAR inhibitors were Myricitrin and Isoarborinol acetate, respectively.
Acknowledgements
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Authors' Contributions
All authors contributed to data analysis, drafting, and revising of the manuscript and agreed to be responsible for all the aspects of this work.
Conflict of Interest
No potential conflict of interest was declared by the authors.
Orcid:
Nurmawati Fatimah: https://www.orcid.org/0000-0002-9661-8934
Arifa Mustika: https://www.orcid.org/0000-0001-6461-5782
Sri Agus Sudjarwo: https://www.orcid.org/0000-0002-7998-7500
Nurul Shahfiza Noor: https://www.orcid.org/0000-0002-5219-3081
--------------------------------------------------------------------------------------
How to cite this article: Nurmawati Fatimah, Arifa Mustika*, Sri Agus Sudjarwo, Nurul Shahfiza Noor, Metabolite profiling based on UPLC-QTOF-MS/MS and evaluation of petiveria alliacea leaves extract as an in silico anti-inflammatory. Journal of Medicinal and Pharmaceutical Chemistry Research, 2024, 6(3), 344-361. Link: http://jmpcr.samipubco.com/article_185013.html
--------------------------------------------------------------------------------------
Copyright © 2024 by SPC (Sami Publishing Company) + is an open access article distributed under the Creative Commons Attribution License(CC BY) license (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
