Document Type : Review Article
Authors
- Rahadian Zainul 1, 2
- Arif Nur Muhammad Ansori 3, 4, 5, 6, 7
- Ahmad Affan Ali Murtadlo 4, 7, 8
- Teguh Hari Sucipto 9
- Viol Dhea Kharisma 4, 7, 8
- Muhammad Hermawan Widyananda 4, 10
- Bayyinatul Muchtaromah 11
- Amaq Fadholly 12, 13
- Muhammad Khaliim Jati Kusala 13
- Vikash Jakhmola 5
- Maksim Rebezov 14, 15
- Sukma Sahadewa 16
- Putu Angga Wiradana 17
1 Department of Chemistry, Faculty of Mathematics and Natural Sciences, Universitas Negeri Padang, Padang, Indonesia
2 Center for Advanced Material Processing, Artificial Intelligence, and Biophysic Informatics (CAMPBIOTICS), Universitas Negeri Padang, Padang, Indonesia
3 Postgraduate School, Universitas Airlangga, Surabaya, Indonesia
4 Generasi Biologi Indonesia Foundation, Gresik, Indonesia
5 Uttaranchal Institute of Pharmaceutical Sciences, Uttaranchal University, Dehradun, India
6 European Virus Bioinformatics Center, Jena, Germany
7 Division of Research and Development, Jalan Tengah, Pasuruan, Indonesia
8 Department of Biology, Faculty of Science and Technology, Universitas Airlangga, Surabaya, Indonesia
9 Dengue Study Group, Institute of Tropical Disease, Universitas Airlangga, Surabaya, Indonesia
10 Department of Biology, Faculty of Mathematics and Natural Sciences, Universitas Brawijaya, Malang, Indonesia
11 Master Program of Biology, Universitas Islam Negeri Maulana Malik Ibrahim, Malang, Indonesia
12 School of Veterinary Medicine and Biomedical Sciences, IPB University, Bogor, Indonesia
13 Research Center for Veterinary Sciences, National Research and Innovation Agency, Bogor, Indonesia
14 Department of Scientific Research, Ural State Agrarian University, Yekaterinburg, Russian Federation
15 Department of Scientific Research, V. M. Gorbatov Federal Research Center for Food Systems, Moscow, Russian Federation
16 Faculty of Medicine, Universitas Wijaya Kusuma Surabaya, Surabaya, Indonesia
17 Research Group of Biological Health, Study Program of Biology, Faculty of Health and Science, Universitas Dhyana Pura, Bali, Indonesia
Abstract
Dengue fever, caused by the dengue virus, is a significant global health concern, with millions of cases reported annually. The absence of specific antiviral treatments or effective preventive measures has fuelled the urgent need for a dengue vaccine. This provides a comprehensive review of the development of a dengue vaccine, summarizing the major milestones, challenges encountered, and recent advancements in this field. This study explores various vaccine strategies employed, including live attenuated vaccines, inactivated vaccines, recombinant subunit vaccines, and viral vector-based vaccines. In addition, the manuscript discusses the clinical trials conducted to evaluate the safety and efficacy of these vaccine candidates.
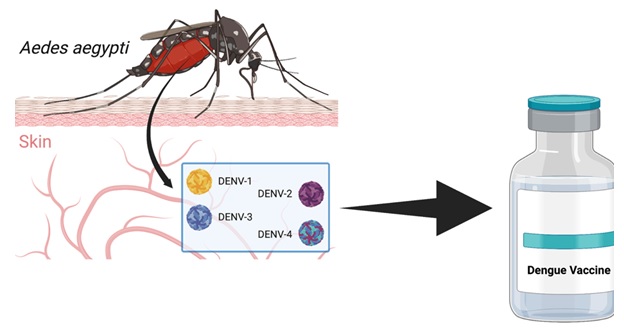
Graphical Abstract
Keywords
Main Subjects
Introduction
The history of dengue fever, a vector-borne disease caused by the dengue virus and transmitted to human through the bite of infected Aedes mosquitoes, is marked by global epidemic outbreaks and substantial morbidity and mortality rates in tropical and subtropical regions [1-5]. According to the World Health Organization (WHO), there are approximately 390 million dengue infections worldwide each year; this may trigger the development of a vaccine to prevent the spread of virus infection [6]. The development of a safe, affordable, and efficacious dengue vaccine is a public health priority that has been long overdue. The journey toward creating a viable vaccine has been fraught with challenges due to the complexity of dengue virus and the risk of antibody-dependent enhancement (ADE) that leads to severe dengue [7].
Dengue virus classification generally consists of four serotypes namely DENV-1, DENV-2, DENV-3, and DENV-4, each capable of causing the full spectrum of disease severity. This complexity has posed significant hurdles for vaccine development. The ideal dengue vaccine should confer protection against all four serotypes simultaneously to prevent ADE. ADE occurs when pre-existing non-neutralizing antibodies from a previous infection with one dengue serotype enhance the entry of a different serotype into host cells, increasing viral replication and potentially leading to severe disease [8,9].
The first vaccine for dengue, Dengvaxia (CYD-TDV) developed by Sanofi Pasteur, was licensed in 2015. This tetravalent live-attenuated vaccine is based on a yellow fever 17D vaccine backbone and has been recommended by the WHO for use in certain endemic regions in individuals aged 9-45 years with evidence of a prior dengue infection. However, this vaccine has faced challenges, including less than optimal efficacy, particularly against DENV-2, and increased risk in previously uninfected (seronegative) people possibly developing severe dengue disease [10,11].
Further efforts to develop a more effective and safer vaccine have led to the exploration of different vaccine platforms, such as subunit, DNA, mRNA, viral vector, and inactivated virus vaccines. Notably, the Takeda’s TAK-003, a live-attenuated tetravalent dengue vaccine, has shown promise in Phase III trials with better efficacy and safety profile compared to Dengvaxia. However, long-term data on its protective immunity are still awaited [12-14].
The development of a dengue vaccine has been shaped by a range of factors, including scientific and technical challenges, ethical considerations, and market dynamics. Regulatory aspects, such as the approval process and post-marketing surveillance, also play a crucial role. It is necessary to comprehend these intricacies to understand the current state of dengue vaccine development and what the future may hold [15].
As the dengue pandemic continues to expand in geographic range and intensity, the quest for an effective vaccine has become even more crucial. It is necessary to understand the past challenges, current advancements, and future directions in dengue vaccine development. This review aims to provide an overview of the progress in dengue vaccine development, discussing the different vaccine platforms, the role of regulatory and ethical considerations, and prospects for future vaccines [16,17].
Structure of dengue virus
The dengue virus belongs to the Flavivirus genus and has a complex structure consisting of three main components: the viral envelope, the capsid, and the viral RNA genome. Understanding the structure of the dengue virus is essential for developing effective vaccines and antiviral therapies [1-3].
Dengue virus has an envelope layer composed of glycoprotein (E) and membrane (M). The E protein plays a crucial role in viral entry into host cells and is the main target for neutralizing antibodies. It is responsible for mediating viral attachment, membrane fusion, and host cell receptor recognition [18,19].
The M protein lies beneath the E protein and is involved in the assembly and budding of viral particles. It also plays a role in the virus's interaction with the host immune system. The E and M proteins together form the outer shell of the virus, giving it its characteristic spherical shape [18,19].
The viral capsid, or core, is located beneath the viral envelope and encloses the viral RNA genome. The capsid protein (C) is responsible for maintaining the structural integrity of viral particle. It also assists in viral replication and packaging of the viral RNA during assembly. The dengue virus genome is a single-stranded positive-sense RNA molecule approximately 11 kilobases in length. It serves as the blueprint for viral replication and protein synthesis. The RNA genome is organized into a single open reading frame (ORF) flanked by untranslated regions (UTRs) at the 5' and 3' ends. The ORF encodes three structural proteins (C, E, M) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5). The non-structural proteins are involved in various aspects of viral replication, assembly, and immune evasion [18,19].
The dengue virus exists as four distinct serotypes, DENV-1, DENV-2, DENV-3, and DENV-4, which share approximately 60-75% genetic similarity. The serotypes differ in their antigenic properties, allowing for the possibility of sequential infections with different serotypes. This phenomenon, known as serotype-specific immunity, plays a crucial role in the development of dengue vaccines [20,21].
Structural studies, including X-ray crystallography and cryo-electron microscopy, have provided valuable insights into the three-dimensional structure of dengue virus. These studies have elucidated the arrangement of the viral proteins and their interactions with host receptors and antibodies, aiding in the development of antiviral therapies and vaccine candidates [22,23].
The structural information has also facilitated the design of recombinant subunit vaccines that utilize specific domains of the E protein to elicit an immune response. EDIII domain on protein E can trigger antibody production with stronger neutralization properties. Recombinant EDIII-based vaccines have shown promise in preclinical and early clinical studies [24].
The structural complexity of dengue virus poses challenges in vaccine development. The high genetic similarity between serotypes requires the vaccines development that can provide simultaneous and balanced protection against all four serotypes. The structural variation between serotypes further complicates the design of broadly protective vaccines [25].
The structure of the dengue virus is a dynamic and intricate assembly of viral proteins and RNA. Understanding the structural components and their functions is crucial for the development of effective vaccines and antiviral strategies against dengue. Continued research into the structural biology of dengue virus will contribute to our understanding of the virus-host interactions and aid in the development of interventions to control and prevent dengue infections [26].
Genome of dengue virus
The genome of dengue virus is a single-stranded positive-sense RNA molecule that serves as the genetic blueprint for viral replication and protein synthesis. Understanding the structure and organization of the dengue virus genome is essential for unravelling its replication mechanisms, genetic diversity, and the development of effective diagnostic tools and antiviral strategies [27].
The dengue virus genome is approximately 11 kilobases in length and contains a single open reading frame (ORF) flanked by untranslated regions (UTRs) at the 5' and 3' ends. The ORF encodes a polyprotein that is subsequently cleaved into three structural proteins (C, prM/M, E) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (Figure 1). These proteins play critical roles in viral replication, assembly, and immune evasion [27].

FIGURE 1 The genome of dengue virus
The 5' and 3' UTRs are non-coding regions that play essential regulatory roles in viral replication and translation. The 5' UTR contains the viral RNA cap structure, which is essential for translation initiation. The 3' UTR contains conserved sequences important for RNA replication and interacts with viral and host proteins during the replication process [28].
The dengue virus genome exhibits a high degree of genetic variability, both within and between the four serotypes (DENV-1, DENV-2, DENV-3, and DENV-4). This genetic diversity is attributed to the high mutation rate of the RNA-dependent RNA polymerase and the occurrence of recombination events during viral replication. These processes contribute to the emergence of new viral variants and the evolution of viral strains with altered virulence and antigenic properties [29].
Genomic studies have identified conserved RNA secondary structures within the UTRs that play crucial roles in viral replication and translation. These structural elements, known as cis-acting replication elements (CREs), are involved in viral RNA synthesis and interact with viral and host proteins to facilitate replication and translation processes [30].
The non-structural proteins encoded by the dengue virus genome are critical for viral replication and evasion of the host immune response. NS3, for example, has protease, helicase, and RNA triphosphatase activities essential for viral polyprotein processing and RNA replication. NS5 possesses RNA-dependent RNA polymerase activity to drive the viral RNA synthesis process [31].
The dengue virus genome's genetic diversity poses challenges for diagnostic and surveillance efforts. The design of molecular diagnostic assays targeting conserved regions is necessary to ensure accurate detection of all four serotypes. In addition, genomic surveillance allows for monitoring viral evolution, identification of emerging strains, and detection of potential vaccine escape mutants [32].
The genome of the dengue virus is also a target for the development of antiviral strategies. Small molecule inhibitors targeting viral enzymes, such as NS3 protease and NS5 polymerase, have shown promise in inhibiting viral replication. Moreover, RNA interference (RNAi) approaches targeting viral RNA have demonstrated antiviral activity against dengue virus in experimental settings [33].
Advancements in next-generation sequencing technologies have facilitated comprehensive genomic studies of dengue virus isolates from different geographic regions. These studies have enhanced our understanding of viral diversity, transmission dynamics, and identification of molecular determinants of viral pathogenesis. This knowledge contributes to the development of more effective vaccines and antiviral therapies [34].
The genome of dengue virus is a single-stranded positive-sense RNA molecule that encodes structural and nonstructural proteins essential for viral replication and evasion of the host immune response. The genetic diversity of the dengue virus poses challenges for diagnosis, surveillance, and control efforts. However, the development of genomic technology has led to a bright idea to study the evolution and pathogenesis of the virus, and facilitate the development of future strategies against dengue.
Serotypes and Variants of Dengue Virus
Dengue virus exists as four distinct serotypes: DENV-1, DENV-2, DENV-3, and DENV-4 (Figure 2). Each serotype shares a high degree of genetic similarity within its own serotype but exhibits significant genetic divergence between serotypes. Understanding the serotypes and variants of dengue virus is crucial for disease surveillance, vaccine development, and understanding the dengue epidemiology [1-3].
Within each serotype, multiple genetic variants or genotypes have been identified. These variants are classified based on genetic differences, particularly within the envelope (E) gene, which encodes the major antigenic determinants. The genetic diversity of dengue virus is driven by a combination of evolutionary forces, including mutation, recombination, and selection pressure from the host immune response[35].

FIGURE 2 Four serotypes of dengue virus
Genetic variants within each serotype can exhibit differences in antigenicity, virulence, and epidemic potential. For example, DENV-2 has been associated with more severe disease outcomes compared to other serotypes, and specific genetic variants within DENV-2 have been linked to outbreaks of severe dengue [36]. Similarly, certain DENV-1 genotypes have been associated with increased epidemic potential and higher transmission rates [37].
The identification and tracking of dengue virus serotypes and variants are essential for disease surveillance. This enables the alterations tracking in circulating strains, detection of new introductions, and prediction of potential disease outbreaks. Genomic surveillance using next-generation sequencing technologies has revolutionized the field by providing detailed insights into the genetic diversity and dynamics of dengue virus populations [38].
The genetic diversity of dengue virus poses challenges for vaccine development. A dengue vaccine must provide protection against all four serotypes to be effective. However, the differences between serotypes and the potential for immune enhancement between serotypes add complexity to vaccine design. Vaccines must elicit a balanced immune response that provides protection against all serotypes while avoiding immune enhancement [39].
Vaccine development efforts have focused on designing tetravalent vaccines that induce immune responses against all four dengue serotypes simultaneously. However, the differences in antigenicity and genetic diversity between serotypes make it challenging to achieve balanced protection. Consequently, vaccine candidates may show differential efficacy against different serotypes, which highlights the need for careful evaluation and selection of vaccine candidates based on their immunogenicity profiles [40].
The identification of variant-specific neutralizing antibodies has opened up new possibilities for the development of serotype-specific vaccines. These vaccines could target specific genetic variants or genotypes within a serotype, providing enhanced protection against specific strains of dengue virus. However, the development and implementation of such variant-specific vaccines present logistical and cost considerations [41].
RNA replication of dengue virus
The RNA replication of dengue virus is a complex process essential for the production of viral progeny and the establishment of infection within host cells. Understanding the mechanisms and key players involved in dengue virus RNA replication is crucial for the development of antiviral strategies and identification of potential drug targets.
The replication of dengue virus RNA genome occurs within specialized replication complexes that are formed within the host cell cytoplasm. These replication complexes are comprised of viral non-structural proteins (NS proteins) and host factors. The NS proteins, particularly NS3 and NS5, play central roles in viral RNA replication[42].
The NS3 protein possesses multiple enzymatic activities, including protease, helicase, and triphosphatase, which are critical for the processing of the viral polyprotein and the unwinding of RNA secondary structures. NS5 is a multifunctional protein with RNA-dependent RNA polymerase activity responsible for the synthesis of new viral RNA molecules [43].
The RNA replication process is initiated by the synthesis of a complementary negative-sense RNA strand, which serves as the template for the production of positive-sense RNA genomes. This step is catalysed by the NS5 RNA-dependent RNA polymerase activity. The negative-sense RNA strand is synthesized using the positive-sense RNA genome as the template [44].
The replication of dengue virus RNA is guided by conserved RNA structures within the 5' and 3' untranslated regions (UTRs) of the viral genome. These structures, known as cis-acting replication elements (CREs), interact with viral and host proteins to facilitate the initiation and progression of RNA replication. The UTRs also contain signals for RNA synthesis initiation and termination [45].
Host factors are crucial for dengue virus RNA replication. Various host proteins are recruited to the viral replication complexes, including those involved in RNA metabolism, translation, and membrane rearrangements. The interactions between viral and host proteins modulate the efficiency and fidelity of viral RNA replication [46].
The process of dengue virus RNA replication occurs in close association with endoplasmic reticulum (ER)-derived membrane components. These membrane rearrangements provide a platform for the assembly of viral replication complexes and protect the viral RNA from host immune surveillance. The viral NS proteins induce the formation of ER-derived structures, which serve as replication sites [47].
The RNA replication of dengue virus is tightly regulated to ensure the balance between viral replication and host immune response evasion. This includes the suppression of host innate immune signalling pathways and the modulation of host translation machinery. NS5 protein has been shown to inhibit the interferon response and interfere with host protein synthesis [48].
Advancements in molecular virology techniques, such as RNA interference (RNAi), have facilitated the identification of host factors involved in dengue virus RNA replication. Genome-wide siRNA screens have revealed host proteins that are essential for viral replication, and targeting these host factors may offer potential antiviral strategies against dengue [49].
Genetic diversity of dengue virus
Dengue virus exhibits a high degree of genetic diversity, both within and between the four serotypes (DENV-1, DENV-2, DENV-3, and DENV-4). This genetic diversity has significant implications for the epidemiology, pathogenesis, and vaccine development of dengue. Understanding the dengue virus's genomic diversity is crucial for effective disease surveillance, prevention, and control strategies. Genetic diversity within each serotype is driven by various evolutionary processes, including mutation, recombination, and selection pressure. These processes contribute to the emergence of different lineages or genotypes within a serotype. Genetic diversity can result in differences in viral fitness, disease severity, and epidemic potential [50].
The envelope (E) gene, which encodes the major antigenic determinants of the virus, exhibits considerable genetic variability. This variability contributes to the ability of dengue virus to escape host immune responses and leads to the emergence of new antigenic variants. Changes in the E gene can affect viral infectivity, virulence, and antibody recognition [51].
Recombination events also play a significant role in generating genetic diversity within dengue virus. Recombination can occur between different serotypes or within the same serotype. Recombinant viruses can acquire genetic material from multiple parental strains, leading to the emergence of novel variants with altered phenotypes. Recombination events can affect viral fitness, disease severity, and transmissibility [52]. Dengue virus genetic variation has significant effects on the dynamics of illness transmission and epidemiology. Different viral lineages or genotypes may exhibit variations in geographic distribution and temporal patterns. Understanding the spatial and temporal distribution of dengue virus variants can help identify high-risk areas, track the spread of outbreaks, and guide public health interventions [53].
Furthermore, genetic diversity impacts the development of dengue vaccines. A successful dengue vaccine needs to provide protection against all four serotypes. However, the genetic diversity within each serotype poses challenges in achieving broad and balanced protection. Vaccines must elicit immune responses that are effective against the diverse array of strains within each serotype [54].
The genetic diversity of dengue virus also affects the accuracy of diagnostic assays. Molecular-based diagnostic tests, such as polymerase chain reaction (PCR) assays, rely on the detection of conserved regions within the viral genome. However, the genetic variability of dengue virus can lead to mismatches between primers/probes and circulating strains, impacting the sensitivity and specificity of diagnostic tests [55].
Next-generation sequencing technologies have revolutionized the study of dengue virus genetic diversity. These techniques allow for high-throughput sequencing of viral genomes, providing comprehensive information on the genetic makeup of viral populations. Genomic studies have revealed the co-circulation of multiple lineages and the presence of viral subpopulations within a given serotype [56].
Phylogenetic analyses of dengue virus sequences have facilitated the classification of strains into distinct lineages and the tracking of viral evolution. These analyses have helped identify the origins of emerging strains, elucidate transmission patterns, and provide insights into the genetic determinants of viral fitness and virulence [57].
The genetic diversity of dengue virus poses challenges for vaccine efficacy and surveillance efforts. Continuous monitoring of viral strains is essential to detect the emergence of new variants that may impact vaccine effectiveness or diagnostic accuracy. Comprehensive genomic surveillance can guide vaccine strain selection, identify vaccine escape mutants, and inform public health strategies for dengue control [58].
Dengue virus in Indonesia
Dengue fever is a significant public health concern in Indonesia, with the country experiencing a high burden of dengue virus transmission. Indonesia is endemic to dengue, and the incidence of dengue fever remains a persistent challenge. Given the presence of all four dengue virus strains, the country faces a complex epidemiological landscape [59].
The prevalence of dengue fever in Indonesia is influenced by several factors, including environmental conditions, population density, and socioeconomic factors. The Aedes aegypti mosquito, the primary vector for dengue transmission, thrives in urban and peri-urban areas, leading to a higher risk of transmission in densely populated regions. The incidence of dengue in Indonesia fluctuates throughout the year, with peaks during the rainy season when mosquito breeding sites increase [60].
Dengue fever poses a substantial burden on the Indonesian healthcare system. The number of reported cases has been steadily increasing over the years, with periodic outbreaks occurring in different regions. Severe forms of the disease, such as dengue haemorrhagic fever (DHF) and dengue shock syndrome (DSS), contribute to significant morbidity and mortality, particularly among children [61].
Several risk factors contribute to the increased transmission of dengue virus in Indonesia. Rapid urbanization and inadequate waste management create conducive breeding sites for Aedes mosquitoes. Climate variability, including temperature and rainfall patterns, can influence the abundance of mosquito populations. Socioeconomic factors, such as housing conditions and access to healthcare, also impact the risk of dengue infection [62].
Efforts to control dengue virus transmission in Indonesia face various challenges. Limited resources for vector control programs, inadequate infrastructure for surveillance and early detection, and low awareness of preventive measures among the population hinder effective control strategies. In addition, the complexity of dengue transmission and diversity of circulating virus strains pose challenges in vaccine development and implementation [63].
The Indonesian government has implemented several strategies to combat dengue fever. These include vector control measures, such as larval source reduction, insecticide spraying, and community-based campaigns promoting the use of mosquito nets and repellents. Health education programs and public awareness campaigns aim to educate the population about dengue prevention and early recognition of symptoms [64].
The introduction of dengue vaccination in Indonesia has the potential to contribute significantly to dengue control efforts. However, the implementation of a dengue vaccine program requires careful consideration of factors such as vaccine effectiveness, cost-effectiveness, and the vaccination integration with existing vector control strategies. Vaccination programs targeting high-risk populations and endemic areas can have a significant impact on reducing dengue burden [65].
Research and collaboration among local and international partners are essential for advancing dengue control in Indonesia. Surveillance studies to monitor dengue virus circulation, clinical trials to evaluate vaccine candidates in local populations, and operational research to optimize control strategies based on local epidemiological data are crucial for guiding evidence-based interventions [66].
Addressing the dengue virus burden in Indonesia requires a comprehensive approach that integrates vector control measures, public awareness campaigns, and potentially dengue vaccination programs. Continued efforts to strengthen surveillance systems, enhance healthcare infrastructure, and promote community engagement are crucial for reducing the impact of dengue fever on the Indonesian population. By implementing a multi-faceted approach and fostering collaborations, Indonesia can work towards mitigating the burden of dengue and improving public health outcomes in the country [67].
Vaccine Strategies for Dengue
Live-attenuated vaccines
Live attenuated vaccines have been a prominent approach in dengue vaccine development. One such vaccine is Dengvaxia® (CYD-TDV), a chimeric tetravalent vaccine developed by Sanofi Pasteur. It incorporates the structural proteins of dengue viruses into the yellow fever vaccine backbone. Clinical trials have shown variable efficacy across different populations, with lower effectiveness observed in seronegative individuals. Potential risk of severe dengue virus infection in seronegative individuals when vaccination is associated with this vaccine.
Live attenuated vaccines replicate in the host cells but have reduced virulence compared to wild-type viruses. Upon vaccination, the attenuated virus enters host cells and triggers an immune response. This immune response includes the production of neutralizing antibodies and the activation of T cells, which recognize and target the viral antigens. The immune response generated by the live attenuated vaccine provides protection against subsequent dengue virus infections [68].
Inactivated vaccines
Inactivated vaccines, either using whole-virus particles or purified antigens, have been explored as an alternative approach. These vaccines offer potential advantages in terms of safety and standardized production processes. However, they have shown limited immunogenicity and efficacy in clinical trials, requiring the use of adjuvants or multiple doses to enhance immune responses [69].
Inactivated vaccines elicit an immune response by presenting the viral antigens to the immune system. These vaccines primarily induce the production of neutralizing antibodies against the viral proteins. The immune response may also include the activation of T cells, contributing to cellular immunity. However, compared to live attenuated vaccines, inactivated vaccines generally induce a weaker cellular immune response [70].
Subunit recombination vaccine technology
Development of subunit recombination vaccine technology against dengue virus through expression of viral proteins or subunits in recombinant expression systems, such as yeast, bacteria, or mammalian cells. The selected proteins or subunits are typically those known to induce a strong immune response and offer protection against dengue infection [71].
Recombinant subunit vaccines induce an immune response by presenting specific viral antigens to the immune system. These antigens often include the envelope (E) and non-structural protein 1 (NS1) of dengue virus. The immune response primarily involves the production of neutralizing antibodies that target the viral antigens and prevent viral entry into host cells. Some recombinant subunit vaccines may also induce cellular immune responses, although to a lesser extent than live attenuated vaccines [71].
Recombinant subunit vaccines have shown variable efficacy in clinical trials. Although they can trigger neutralization activity of antibodies to specific dengue virus serotypes, the coverage level may vary. Enhancing the immunogenicity and breadth of protection is an ongoing challenge for subunit vaccines [72].
Viral vector-based vaccines
Viral vector-based vaccines against dengue virus are developed by genetically modifying non-pathogenic viruses to express dengue viral antigens. Commonly used vectors include adenoviruses, vesicular stomatitis viruses (VSV), alphaviruses, and lentiviruses. The modified viral vectors act as delivery vehicles to introduce the dengue viral antigens into host cells [73].
Viral vector-based vaccines work by infecting host cells with the modified viral vector, which then produces and presents dengue viral antigens to the immune system. This leads to the activation of both humoral and cellular immune responses. The immune response generated by the viral vector-based vaccines provides protection against subsequent dengue virus infections [73].
Viral vector-based vaccines induce robust immune responses, including the production of neutralizing antibodies and the activation of cellular immune responses. The presentation of viral antigens by the viral vectors enhances the recognition and targeting of the antigens by the immune system. These vaccines have demonstrated the ability to elicit strong and durable immune responses against dengue virus [73].
Each vaccine strategy presents its own advantages and challenges in dengue vaccine development. Balancing safety, immunogenicity, and efficacy is crucial for the successful development of a dengue vaccine. Further research is needed to optimize vaccine formulations, adjuvants, and delivery systems to achieve optimal immune responses and long-term protection [74].
Furthermore, considerations such as vaccine dosing regimens, timing of vaccination, and age-specific responses need to be explored to ensure maximum effectiveness. The development of a safe and effective dengue vaccine that provides long-lasting immunity against all four serotypes remains a significant global health priority [74].
Clinical Trials and Regulatory Considerations
Clinical trials for dengue vaccines
Clinical trials play a crucial role in assessing the safety, immunogenicity, and efficacy of dengue vaccine candidates. These trials typically follow a phased approach, starting with Phase 1 studies to evaluate safety and dosing in a small group of healthy volunteers. Phase 2 trials expand the sample size and assess immunogenicity and dosage optimization. Phase 3 trials involve larger populations and evaluate vaccine efficacy and safety in endemic areas with diverse dengue serotypes [75].
Immunogenicity and efficacy endpoints
Immunogenicity endpoints in dengue vaccine trials often include the measurement of neutralizing antibody titers, as well as cell-mediated immune responses. Efficacy endpoints focus on the prevention of symptomatic dengue cases or the reduction of severe disease, hospitalizations, and deaths. Vaccine efficacy is determined by comparing the incidence of dengue in vaccinated and placebo/control groups over a defined period [76].
Serostatus and prior dengue exposure
Prior dengue exposure and serostatus can significantly impact vaccine effectiveness. Individuals with prior exposure to one or more dengue serotypes typically exhibit higher immune responses upon vaccination. However, there are conditions where an individual who is seronegative to dengue infection may have an increased risk of being infected with a different serotype. Careful consideration of serostatus and prior dengue exposure is crucial in determining vaccine safety and efficacy in different target populations [77].
Regulatory considerations
Regulatory authorities play a vital role in evaluating the safety and efficacy of dengue vaccine candidates. Stringent regulatory processes ensure that vaccines meet the necessary quality, safety, and efficacy standards before approval for public use. Regulatory agencies assess clinical trial data, including immunogenicity, efficacy, and safety outcomes, to make informed decisions regarding licensure and recommendations for vaccine use [78].
Post-licensure surveillance and long-term safety
Post-licensure surveillance is crucial for monitoring the long-term safety and effectiveness of dengue vaccines in real-world settings. It allows for the detection of rare adverse events and provides ongoing data on vaccine performance, including duration of protection and waning immunity. Robust surveillance systems facilitate the early detection and management of any potential safety concerns [79].
Vaccination strategies and implementation
The introduction of a dengue vaccine into immunization programs requires careful consideration of factors such as target populations, age groups, and optimal vaccination strategies. Integrated vector control measures, public awareness campaigns, and sustainable vaccine delivery systems are essential components of successful dengue control programs. Comprehensive understanding of the clinical trial process, regulatory considerations, and post-licensure surveillance is critical for the successful development, evaluation, and implementation of dengue vaccines. Continued collaboration between researchers, regulatory agencies, and public health authorities is key to advancing dengue vaccine development and ultimately reducing the global burden of dengue fever [80].
Challenges and Future Perspectives
One of the main challenges in dengue vaccine development is the complexity of the dengue virus itself. Infection with one of the four dengue virus serotypes, DENV-1, DENV-2, DENV-3, and DENV-4, does not confer long-term immunity or against infection with other serotypes. This increases the risk of severe dengue during subsequent infections, a phenomenon known as antibody-dependent enhancement (ADE). ADE complicates vaccine development as vaccines must be tetravalent, providing balanced and lasting immunity against all four serotypes simultaneously [81-83].
Secondly, the variability of immune response among individuals, influenced by factors such as age, previous exposure to the virus, and co-infections, further complicates the development and evaluation of a dengue vaccine. This was highlighted by the issue with Dengvaxia, where it was found to increase the risk of severe dengue in those who had not been previously exposed to the virus (seronegative individuals) [84,85].
The interpretation of clinical trial results presents another challenge. The endpoints used to evaluate vaccine efficacy in clinical trials can be influenced by a multitude of factors, including the level of dengue virus transmission in the community, the age of the participants, and their immune status at the time of vaccination. There is an ongoing debate over what constitutes a robust measure of vaccine efficacy [86].
Further challenges include the logistical and economic factors of vaccine distribution and administration, particularly in resource-limited settings. Vaccine implementation strategies need to account for the cost-effectiveness, accessibility, and the cold chain infrastructure for vaccine storage and transportation. Vaccination campaigns also need to ensure high coverage to achieve herd immunity [87].
On the regulatory front, ensuring the ethical conduct of clinical trials, particularly in resource-limited, dengue-endemic countries, poses a significant challenge. Vaccine trials should adhere to the highest ethical standards, which include informed consent, ensuring a favourable risk-benefit ratio for participants, and post-trial access to the vaccine [88].
Looking forward, newer vaccine platforms such as mRNA and viral vector-based vaccines offer new possibilities for dengue vaccine development. The success of mRNA vaccines for COVID-19 provides an optimistic outlook for this technology, which offers advantages in terms of safety, speed of manufacturing, and flexibility in changing the encoded antigen [89,90].
Innovative trial designs and statistical methods could also help overcome some of the challenges in evaluating vaccine efficacy. Composite endpoints, long-term follow-up to evaluate duration of protection, and multi-regional trial designs could provide more robust measures of efficacy [91-93].
There is also a need for more detailed understanding of the immune response to dengue infection and vaccination, including the mechanisms behind ADE. This could help in the development of new vaccine candidates and immunization strategies, and in predicting vaccine performance [92,94].
Public-private partnerships could play a pivotal role in overcoming the logistical and economic challenges of vaccine implementation. Governments, international organizations, vaccine manufacturers, and NGOs need to collaborate to ensure the accessibility and affordability of dengue vaccines, particularly in resource-limited settings[95,96].
Conclusion
The development of a safe, effective, and affordable dengue vaccine is crucial for combating dengue fever. Advancements in dengue immunology have led to various vaccine strategies, including live attenuated, inactivated, recombinant subunit, and viral vector-based vaccines. However, challenges remain, such as achieving balanced protection against all four serotypes, mitigating immune enhancement risks, ensuring long-term durability, and addressing cost and access issues. Global collaboration among researchers, regulatory authorities, policymakers, and public health organizations is crucial for progress. Integrating vaccination programs with comprehensive prevention and control strategies is essential for maximum impact.
Acknowledgements
The authors would like to thank Jalan Tengah, Indonesia (https://jalantengah.site) for editing the manuscript.
ORCID
Rahadian Zainul*: https://www.orcid.org/0000-0002-3740-3597
Arif Nur Muhammad Ansori: https://www.orcid.org/0000-0002-1279-3904
Ahmad Affan Ali Murtadlo: https://www.orcid.org/0000-0002-7942-875X
Teguh Hari Sucipto: https://www.orcid.org/0000-0003-0512-2990
Viol Dhea Kharisma: https://www.orcid.org/0000-0001-9060-0429
Muhammad Hermawan Widyananda: https://www.orcid.org/0000-0002-0064-8865
Bayyinatul Muchtaromah: https://www.orcid.org/0000-0001-9968-8295
Amaq Fadholly: https://www.orcid.org/0000-0003-2064-8552
Muhammad Khaliim Jati Kusala: https://www.orcid.org/0000-0002-7613-721X
Vikash Jakhmola: https://www.orcid.org/0000-0002-8108-006x
Maksim Rebezov: https://www.orcid.org/0000-0003-0857-5143
Sukma Sahadewa: https://www.orcid.org/0009-0009-9253-7633
Putu Angga Wiradana: https://www.orcid.org/0000-0002-0139-8781
-----------------------------------------------------------------------
How to cite this article: Rahadian Zainul*, Arif Nur Muhammad Ansori, Ahmad Affan Ali Murtadlo, Teguh Hari Sucipto, Viol Dhea Kharisma, Muhammad Hermawan Widyananda, Ayyinatul Muchtaromah, Amaq Fadholly, Muhammad Khaliim Jati Kusala, Vikash Jakhmola, Maksim Rebezov, Sukma Sahadewa, Putu Angga Wiradana, The recent development of dengue vaccine: a review. Journal of Medicinal and Pharmaceutical Chemistry Research , 2024, 6(4), 362-382. Link: http://jmpcr.samipubco.com/article_184982.html
-----------------------------------------------------------------------
Copyright © 2024 by SPC (Sami Publishing Company) + is an open access article distributed under the Creative Commons Attribution License(CC BY) license (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.